Analysis of the molecular diversity of R. solani isolates from five Ugandan agro-ecological zones using inter - simple sequence repeat markers
Received: May 30, 2018
Accepted: September 01, 2018
Published: November 01, 2018
Genet.Mol.Res. 17(4): http://dx.doi.org/gmr16039927
DOI: http://dx.doi.org/10.4238/gmr16039927
Abstract
This study used seven inter-simple sequence repeat (ISSR) markers to characterize 55 R. solani isolates in order to establish their genetic diversity. The isolates were from five common bean growing agro-ecological zones of Uganda; the Southwest highlands (SW), Lake Victoria crescent and Mbale farmlands (LVC), Teso farming system (TFS), the Northern mixed farmlands (N) and the Western highlands and Semliki flats (W). DNA was extracted from 7-day old R. solani isolates growing on potato dextrose agar and amplified using PCR. Molecular analysis based on the ISSR fingerprints established moderate genetic diversity (mean = 0.389 ± 0.022). In addition, a high degree of intraspecific variation was observed by cluster analysis which generated a dendrogram with 5 highly reliable clusters (p ≥ 0.95) whose composition was independent of agro-ecological zones. Further evidence of intraspecific variation was provided by analysis of molecular variance which ascribed all the observed variation to genetic differences within the isolates. Isolates from SW, W, TFS and N had significant standardized index of association values indicative of clonal reproduction while isolates from LVC did not have significant values whence were sexual. The genetic diversity revealed by our study means that screening common bean germplasm for sources of resistance to R. solani should be done using diverse pathotypes and secondly, the breeders should pyramid many disease resistance genes in elite cultivars to provide durable resistance against R. solani.
Keywords
Genetic diversity; Analysis of molecular variance; Standardized index of association; Intraspecific variation
Introduction
Common bean (Phaseolus vulgaris L.) is one of the most ancient crops of the new world and it shows remarkable plasticity in terms of cultivation methods, environmental adaptation and morphological variability. Production exceeds 23 million metric tonnes of which 7 million are produced in Latin America and Africa. Per capita consumption in areas near the Great lakes region of Africa has been reported to be as high as 40Kg/year (Blair et al., 2010). The great production figures have largely been achieved because of intensive cropping systems brought about by high population density and shrinking cropping area. However, intensification has led to increasing levels of the inoculum of soil borne pathogens and imbalance between harmful and beneficial microbes.
Rhizoctonia solani Kühn (Teleomorph: Thanatephorus cucumeris (Frank) Donk) is a soil borne pathogen thatcauses a number of diseases in economically important crops. It thrives through mycelial or sclerotial forms in plant debris in the field (Godoy-Lutz et al., 2000) and it exists in either multinucleate or binucleate forms.
Rhizoctonia solani, a member of the multinucleate Rhizoctonia group (Carling, 1996) is divided into 14anastomosis groups (AGs)- based on hyphal anastomosis reactions (Sneh et al., 1991, 1996; Carling et al., 1999, 2002). Binucleate R. solani are grouped from AGA to AGS (Sneh et al., 1991) and are generally non-pathogenic. Although classification on the basis of AGs still remains a valid way of characterizing R. solani, it is time consuming and ambiguous due to the presence of bridging isolates and isolates that have lost the ability to fuse (Hyakumachi and Ui 1987; Sharon et al., 2006). Other studies (Yokoyama and Ogoshi, 1986) showed that nutritional conditions of the growth media affected the fusion reactions.
Consequently, molecular tools have been developed to offset the challenges posed by characterization on the basis of the AGs. Random amplified polymorphic DNA (RAPD) markers, inter- simple sequence repeat (ISSR), simple sequence repeat markers (SSR) and polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) have been used to study the extent of diversity among R. solani (Mwang’Ombe et al., 2007; Kilicoglu and Ozkoc, 2010; Ganeshamoorthi and Dubey, 2013; Dubey et al., 2014;)
In order to accelerate efforts to breed for resistance to R. solani in common bean, it is important to understand the genetic variation that exists between isolates to inform host-pathogen studies. Although the genetic diversity of other root rots pathogens especially Fusarium and Pythium spp. in Uganda has been studied extensively (Tusiime, 2003; Mukalazi, 2004) not much is known about the genetic diversity inherent in Ugandan R. solani isolates. Therefore, the aim of this study was to elucidate the genetic diversity of R. solani using inter -simple sequence repeat markers (ISSR) sometimes called random amplified microsatellites (RAMS). ISSR marker technology involves amplification of a DNA fragment flanked by varying numbers of repeat motifs oriented in opposite directions (Bornet and Branchard, 2001). Elucidation of the genetic diversity of R. solani will not only accelerate the deployment of integrated pest management (IPM) strategies but will also aid bean breeding programs accelerate the development of varieties that are resistant to the pathogen.
Material and Methods
Plant sample collection, pathogen isolation and purification
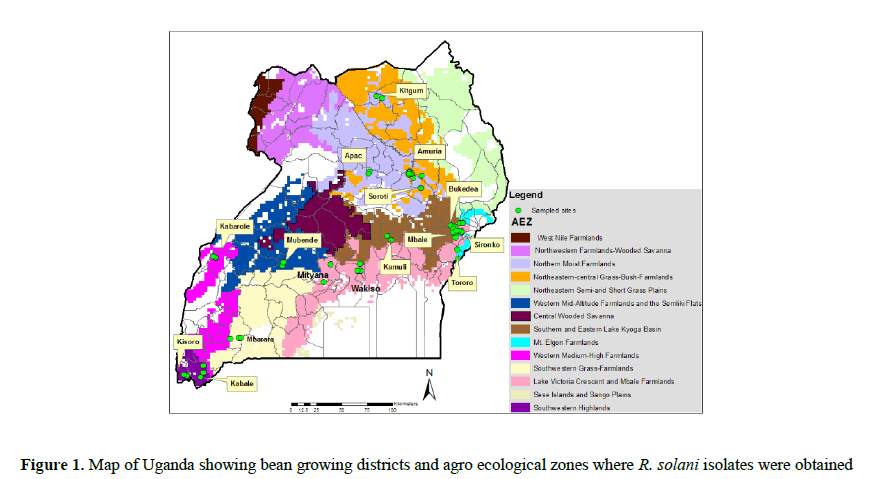
Bean plants showing symptoms of Rhizoctonia root rots were collected from over 300 farmers’ fields in 15 Ugandan districts in five agro-ecological zones (Figure 1); Southwest highlands (SW; annual rainfall >1200 mm, altitude 1247-2313 m), Lake Victoria crescent and Mbale farmlands (LVC; annual rainfall >1215-1328 mm, altitude 1040-1433 m), Teso Farming System Zone (TFS; annual rainfall > <1000 mm, altitude 1072-1130 m), the Northern mixed farmlands (N; (annual rainfall >1197 mm, altitude 942-1182m) and the Western Mixed Farming System (W; annual rainfall >1000-1200 mm, altitude 1052-1501 m) agro-ecological zones (AEZs) during the cropping season of September- October, 2013. Temperature and rainfall data was obtained from a website www.erails.net/UG/aris/kms, while altitude was obtained from GPS readings taken during the surveys.
Isolation of Rhizoctonia spp. was conducted at the International Centre for Tropical Agriculture (CIAT) -Uganda laboratory at the National Agricultural Research Laboratories (NARL)-Kawanda. Infected roots were washed under running tap water, blot dried in tissue napkin in the laminar flow chamber. Small pieces of ~ 5 mm long were cut and placed on water agar (WA) media (13.2 g/ L, Biolife Italiana, Milano, Italia) amended with 0.03g/L Rifamycin antibiotic (Sigma-Aldrich Chemie GmbH, Steinheim, Germany), and incubated for 24 h in darkness. Emerging fungal colonies were then transferred to potato dextrose agar (PDA), [19.5 g/L, Sigma-Aldrich Chemie GmbH, Steinheim, Germany] supplemented with 0.03 g/L Rifamycin antibiotics (Sigma-Aldrich Chemie GmbH, Steinheim, Germany. Following fungal purification using the hyphal tipping method, the isolates were grown on WA, and using a sterile needle under a stereo microscope (Leica, Wild M3B, Leica systems, Germany), and culture tips were grown on PDA amended with 0.03 g/L Rifamycin.
DNA extraction and amplification
Mycelia (150 mg) from 7-day R. solani cultures was harvested and DNA extracted from 55 isolates using the method of Mahuku (2004). The concentration of the DNA was determined using the DyNaQuant DQ300 fluorometer (Hoefer Inc, Holliston, MA) and the concentrations adjusted to 100 ng/µL for use in the amplification reactions. A total of fifteen inter simple sequence repeat (ISSR) markers markers previously used to characterise fungal genomes (Hantula et al., 1996; Abadio et al., 2012) were run on a subset of 10 randomly chosen isolates to identify the most polymorphic ones. Seven polymorphic ISSR markers (Table 1) were then used to characterize the entire population of 55 isolates.
| Primer name | Primer sequence (5’-3’) | T (°C) |
|---|---|---|
| UBC 809 | (AG)8G | 45 |
| UBC 836 | (AG)8YA | 45 |
| UBC 888 | (GA)8 YG | 45 |
| UBC 891 | GGAGAGGAGAGGAGA | 45 |
| RAMS 1 | BDB (CA)7 | 45 |
| RAMS 2 | DBD (AC)6CA | 55 |
| RAMS 3 | BDB (ACA)5 | 50 |
Y= (G, A or C); B = (G, T or C); D = (G, A or T).
Table 1: Annealing temperature (T) and sequences of ISSR markers used to characterise 55 R. solani isolates.
Syngene G: BOX gel documentation system (Syngene, Fredrick, MD). All PCR reactions were carried out twice to ensure the reproducibility of the reactions. Only samples that showed consistent clear banding patterns during the two runs were considered. A binary matrix (0/1) to represent absence/presence of an allele was generated from the banding patterns observed with all the markers. DNA fragments greater than 2000 base pairs were omitted from the scoring because their amplification efficiency was low.
Analysis of molecular diversity
The index of inbreeding (fixation index), defined as FIS= 1- (HO/HE), was calculated over all polymorphic loci, where HO and HE correspond to average observed and expected heterozygosity, respectively. The probability of outcrossing t was estimated using the equilibrium value of FIS, which is defined as (1-t)/ (1+t) or t= (1-FIS) / (1+FIS) (Hartl and Clark, 1987).
A genetic distance matrix based on Jaccard’s index was calculated from the binary matrix. The Jaccard distance was chosen because it does not use the absence of an allele as a shared characteristic (Legendre and Legendre, 1998). Isolates were then clustered by using the unweighted pair grouping by mathematical averaging (UPGMA) algorithm. UPGMA was chosen because it makes no assumptions about evolutionary rate or phylogenetic relationships. Uncertainty for each cluster was assessed by means of p values calculated via 999 multiscale bootstrap resampling repetitions. The P value of a cluster is a value between 0 and 1, indicating how strong the cluster is supported by data. The R package pvclust (Suzuki and Shimodaira, 2014) was used for generating the distance matrix and clustering the data. Nei’s unbiased haploid diversity index (Nei,1987) was calculated using GenAlEx version 6.503 (Peakall and Smouse, 2012).
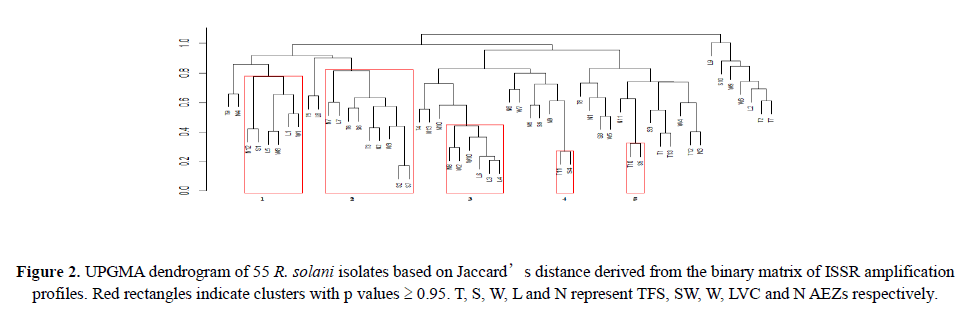
Linkage disequilibrium was quantified by estimating the unbiased index of association (Agapow and Burt, 2001) within populations and among loci using R package poppr v 2.5.0 (Kamvar et al., 2014). The index detects multilocus linkage signifying association between alleles at different loci and clonal reproduction in populations. Significance departure (α=0.05) from the null hypothesis (no linkage among loci) was tested using 2999 permutations. Genetic differentiation within isolates and among AEZs was carried out by performing analysis of molecular variance (AMOVA) using the Manhattan distance generated from the binary matrix based on 999 permutations. The R package vegan (Oksanen et al., 2017) was used for this purpose (Figure 2).
Results
All the seven ISSR markers were very polymorphic generating band sizes ranging from 300-2000 base pairs (Figures 3A-3C). The polymorphism was also evident in the proportion of polymorphic loci. LVC isolates had the greatest proportion of polymorphic loci while isolates from N had the lowest proportion. The primers amplified all the R. solani isolates and the UPGMA dendrogram (Figure 2) based on Jaccard’s distance showed moderate diversity (mean uh=0.389±0.022). Five highly reliable clusters (p ≥ 0.95) comprising of 45% of the isolates were generated. Clusters 1 and 2 consisted of isolates from T and SW while the other clusters had a mixture of isolates from all the AEZs. Generally, the isolates did not cluster according to AEZ.
The isolates showed a mixed mode of reproduction. Isolates from T, N, SW, W had a clonal mode of reproduction while isolates from LVC were sexual. In addition, the isolates not only showed heterozygote excess but were also outbreeding as indicated by the negative values of the fixation index (FIS) and the outbreeding coefficient (t) respectively. All these results are summarized in Table 2.
| AEZ | N | uh | rBarD | Fis | t | P |
|---|---|---|---|---|---|---|
| TFS | 13 | 0.35 (0.023) | 0.009 | -0.128 | 0.77 | 85 |
| N | 13 | 0.34 (0.027) | 0.016 | -0.161 | 0.72 | 77 |
| SW | 10 | 0.42 (0.032) | 0.011 | -0.11 | 0.79 | 76 |
| LVC | 9 | 0.41 (0.036) | 0.006 | -0.18 | 0.68 | 98 |
| W | 10 | 0.39 (0.027) | 0.002 | -0.11 | 0.8 | 82 |
| Total | 55 | 0.39 (0.022) | -0.141 | 0.75 | 84 (4.02) | |
AEZ: agro-ecological zone; N: number of samples; rBarD: unbiased index of association; uh: Nei’s unbiased haploid diversity index = (N/N-1); FIS: inbreeding coefficient estimate; t, outbreeding coefficient estimate; P: percentage of polymorphic loci. Values in parentheses are standard error of the mean (SEM). In column 4, values in bold represent significant p values (α=0.05)
Table 2: Genetic variability, linkage disequilibrium, inbreeding and outbreeding probability values of 55 Ugandan R. solani isolates based on seven ISSR markers
Analysis of molecular variance (AMOVA) attributed all the observed variation to genetic differences within the isolates. AMOVA results are summarized in Table 3.
| Source of variation | df | Sum sq | Mean sq | Variance component | % |
|---|---|---|---|---|---|
| Between AEZ | 4 | 44.290 | 11.07 | 0 | 0 |
| Between districts within AEZ | 13 | 154.060 | 11.85 | 0 | 0 |
| Within districts | 37 | 450.590 | 12.18 | 12.17 | 100 |
| Total | 54 | 648.940 | 12.02 | 12.17 | 100 |
df- Degrees of freedom; Sum sq-sum of squared deviations from the mean; Mean sq-mean squares deviation.
Table 3. Analysis of molecular variance (AMOVA) of 55 Ugandan R. solani isolates showing population differentiation at various strata
Discussion
Fifty-five isolates of R. solani were isolated from common bean plants showing characteristic symptoms and characterized using ISSR markers which showed a high degree of polymorphism. This finding is supported by the total percentage of polymorphic which was >70%. Our findings are much lower than Dubey et al. (2014) who reported 99.7% percent polymorphism when studying the diversity of R. solani isolates affecting different pulse crops in India. This could be due to the different sequences of ISSR markers used in our two studies. The high degree of polymorphism obtained is attributed to the hyper-variable nature of the ISSR markers which enables the detection of a higher level of polymorphism than would be observed with other markers. ISSR markers provide a sensitive, fast and low cost alternative for the fingerprinting of plant pathogenic fungi.
The R. solani isolates showed moderate intraspecific variation. This finding is supported by the results of AMOVA which attributed all the variation to differences within isolates. Similar results have been reported by Gonzalez et al. (2012) who studied 92 isolates of Rhizoctonia solani subgroups AG-1-IE and AG-1-IF causing web blight of common bean and Mwang’Ombe et al. (2007) who studied the diversity of 41 Kenyan R. solani isolates affecting common bean using SSR markers. However, the Kenyan study reported a broader range of the gene diversity index than our study possibly because of the marker type used. SSR markers are co-dominant and are therefore able to detect greater diversity than ISSR markers. Since it has been reported that outbreeding species maintain most of the genetic variation within rather than among populations (James et al.,1999), we think the R. solani population in Uganda is outbreeding. Our conclusion is supported by the high probability of outcrossing, t obtained in this study. The R. solani isolates from the Ugandan AEZs have a single haploid nucleus that combines with another nucleus carrying a compatible mating type to complete their life cycle (heterothallism). In addition, the clustering of the isolates within the clusters which was independent of AEZs also suggests intraspecific variation probably due to mutations which created new phenotypes upon which natural selection acted (Schardl and Craven,2003). Another possible explanation for observed variation could be microevolution. It has been reported (Brasier,1995) that the haploid genetic systems of fungi, their fast-reproductive rates and anastomosis reactions that favor exchange of genetic material between species make fungi prone to microevolution. Although the fast replication of haploid genomes may hasten evolution because of mutations, parasexuality and interspecific mating encourage large scale genomic restructuring. Consequently, the hybrids merge the traits of their parents or acquire increased genetic fitness to colonize new ecological niches. Further, variation could also be due to meiotic recombination that results in the development of basidiospores (Cubeta and Vilgalys, 1997) or the transmission of vegetative mycelia across short distances by rain splash or over long distance by movement of seed (Galvez et al., 1989).
The R. solani isolates showed excess heterozygosity. This finding is supported by the negative values of the inbreeding coefficient, FIS. Similar findings have been reported in R. solani affecting rice by Rosewich et al., (1999) although comparisons would not be convincing because they used RFLP while our study used ISSR markers. A number of mechanisms favoring heterozygosity have been proposed including over dominant selection (Mitton, 1989), associative over dominance (Nei, 1987) and negative assortative mating (Hartl and Clark, 1989). The role of over dominance in our study is not plausible because of the neutral ISSR loci. Since positive assortative mating increases homozygosity due to the mating of like phenotypes (Hartl and Clark, 1989) we believe the observed excess heterozygosity could be a consequence of negative assortative mating.
Conclusion
Our study not only characterized R. solani isolates using ISSR markers but also established the diversity inherent in the pathogen. The genetic diversity revealed by our study has implications for breeders. Firstly, screening common bean germplasm for sources of resistance to R. solani should be done using diverse pathotypes and secondly, the use of resistant cultivars to control R. solani can only be effective if breeders pyramid many disease resistance genes in elite cultivars.
About the Authors
Corresponding Author
A.S. Male
Plant Pathology and Biotechnology Unit, International Centre For Tropical Agriculture, Kampala, Uganda
- Email:
- a.m.ssekamate@cgiar.org
References
- Abadio AKR, Lima SS, Santana MF and Queiroz M (2012) Genetic diversity analysis of isolates of the fungal bean pathogen Pseudocercospora griseola from central and southern Brazil. GMR. 11:1272-1279. https://doi.org/10.4238/2012.may.14.1
- Agapow PM and Burt A (2001) Indices of multilocus linkage disequilibrium. Molecular Ecology Resources. 1:101-102.https://doi.org/10.1046/j.1471-8278.2000.00014.x
- Blair MW, Gonzalez LF, Kimani PM and Butare L (2010) Genetic diversity, inter-gene pool introgression and nutritional quality of common beans (Phaseolus vulgaris L.) from Central Africa. Theor Appl Genet. 121: 237-248. https://doi.org/10.1007/s00122-010-1305-x
- Bornet B and Branchard M (2001) Non-anchored inter simple sequence repeat markers: Reproducible and specific tools for genome fingerprinting. Plant Molecular Biology Reporter. 19: 209-215. https://doi.org/10.1007/bf02772892
- Brasier CM (1995) Episodic selection as a force in fungal microevolution, with special reference to clonal speciation and hybrid introgression. Canadian Journal of Botany. 73: S1213–S1221. https://doi.org/10.1139/b95-381
- Carling DE (1996) First report of powdery scab of potatoes in Alaska. Plant Disease. 80:1208. https://doi.org/10.1094/pd-80-1208b
- Carling DE, Pope EJ, Brainard KA and Carter DA (1999) Characterization of mycorrhizal isolates of Rhizoctonia solani from an orchid, including AG 12, a new anastomosis group. Phytopathology. 89: 942-946. https://doi.org/10.1094/phyto.1999.89.10.942
- Carling DE, Kuninaga S and Brainard KA (2002) Hyphal anastomosis reactions, rDNA-internal transcribed spacer sequences, and virulence levels among subsets of Rhizoctonia solani Anastomosis Group 2 (AG 2) and AG BI. Phytopathology. 92: 43-50. https://doi.org/10.1094/phyto.2002.92.1.43
- Cubeta MA and Vilgalys R (1997) Population biology of the Rhizoctonia solani complex. Phytopathology. 87 (4):480-484.
- Dubey SC, Tripathi A and Upadhyay BK (2014) Molecular diversity analysis of Rhizoctonia solani isolates infecting various pulse crops in different agro-ecological regions of India. World J Microbiol Biotechnology.30 (6): 1699-1715. https://doi.org/10.1007/s12223-012-0165-y
- Galvez GE, Mora B and Pastor-Corrales MA (1989) Web blight. In: Schwartz HF, Pastor-Corrales MA (eds.). Bean production problems in the tropics. CIAT, Cali, pp 195–259.
- Ganeshamoorthi P and Dubey SC (2013) Phylogeny analysis of Indian strains of Rhizoctonia solani isolated from chickpea and development of sequence characterized amplified region (SCAR) marker for detection of the pathogen. African Journal of Microbiology Research. 7: 5516-5525. https://doi.org/10.5897/ajmr2013.5769
- Godoy-Lutz G, Arias J, Steadman JR, Eskridge KM (1996) The web blight pathogen: Its effect on common bean seed quality, germination and early disease development. Bean Improvement Cooperative. 39: 152-153.
- Gonzalez N, Godoy-Lutz G, Steadman JR, Higgins R and Eskridge KM (2012) Assessing genetic diversity in the web blight pathogen Thanatephorus cucumeris (anamorph = Rhizoctonia solani) subgroups AG-1-IE and AG-1-IF with molecular markers. J Gen Plant Pathol. 78:85–98. https://doi.org/10.1007/s10327-012-0361-2
- Hantula J, Dusabenyagasani M and Hamelin RC (1996). Random amplified microsatellites (RAMS)-a novel method for characterizing genetic variation within fungi. Eur J for Path. 26: 159-166.
- Hartl D and Clark A (1997) Principles of population genetics. 3rd edition. Sinauer Associates, Sunderland, Massachusetts, USA.
- Hyakumachi M and Ui T (1987) Non-self-anastomosing isolates of Rhizoctonia solani obtained from fields of sugar-beet monoculture. Transactions of the British Mycological Society. 89: 155-159. https://doi.org/10.1016/s0007-1536 (87)80147-x
- James TY, Porter D, Hamrick JL and Vilgalys R (1999) Evidence for limited intercontinental gene flow in the cosmopolitan mushroom, Schizophyllum commune. Evolution.53: 1665-1677. https://doi.org/10.2307/2640430
- Kamvar ZN, Tabima JF and Grünwald NJ (2014) poppr an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ. 2: e281. https://doi.org/10.7287/peerj.preprints.161
- Kilicoglu MC and Özkoc I (2010) Molecular characterization of Rhizoctonia solani AG4 using PCR-RFLP of the rDNA-ITS region. Turk J Biol.34: 261-269. https://doi.org/10.1016/s1874-5334 (04)80011-x
- Legendre P and Legendre L (1998) Numerical Ecology, Elsevier, Amsterdam, The Netherlands. https://doi.org/10.1016/s0304-3800 (00)00291-x
- Mahuku GS (2004) A simple extraction method suitable for PCR- based analysis of plant, fungal, and bacterial DNA. Plant Molecular Biology Reporter. 22: 71-81. https://doi.org/10.1007/bf02773351
- Mitton JB (1989) Physiological and demographic variation associated with allozyme variation. In: Soltis DE, Soltis PS (eds.). Isozymes in plant biology, Chapman and Hall, London, UK Pp 87-105 https://doi.org/10.1007/978-94-009-1840-5_7
- Mukalazi J (2004) Pathogen variation and quantification of Pythium spp. in bean fields in Uganda. Ph.D. thesis. Makerere University, Kampala, Uganda.
- Mwang’Ombe AW, Thiong’O G, Olubayo FM and Kiprop EK (2007) DNA microsatellite analysis of Kenyan isolates of Rhizoctonia solani from common bean (Phaseolus vulgaris L.). Plant pathology journal. 6: 66-71. https://doi.org/10.3923/ppj.2007.66.71
- Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York, USA. https://doi.org/10.1007/978-1-4684-4988-4
- Oksanen J, Blanchet G, Friendly M, Kindt R, Legendre P et al. (2017) vegan: community ecology package. R package version 2.4-3. https://CRAN.R-project.org/package=vegan
- Peakall R and Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics. 28: 2537-2539. https://doi.org/10.1093/bioinformatics/bts460
- Rosewich UL, Pettway RE, McDonald BA and Kistler HC (1999) High levels of gene flow and heterozygote excess characterize Rhizoctonia solani AG-1 IA (Thanatephorus cucumeris) from Texas. Fungal Genet. Biol. 28: 148-159. https://doi.org/10.1006/fgbi.1999.1174
- Schardl CL, Craven KD (2003) Interspecific hybridization in plant-associated fungi and oomycetes: a review. Molecular Ecology. 12,2861-2873. https://doi.org/10.1046/j.1365-294x.2003.01965.x
- Sharon M, Kuninaga S, Hyakumachi M and Sneh B (2006) The advancing identification and classification of Rhizoctonia spp. using molecular and biotechnological methods compared with the classical anastomosis grouping. Mycoscience. 47: 299-316. https://doi.org/10.1007/s10267-006-0320-x
- Sneh B, Burpee L and Ogoshi A (1991) Identification of Rhizoctonia species. APS press., St Paul, MN, USA.
- Sneh B, Jabaji-Hare S, Neate S and Dijst G (1996) Rhizoctonia species: Taxonomy, molecular biology, ecology, pathology and disease control. 1st ed. Springer-science+ business media., Dordrecht, The Netherlands.
- Suzuki R and Shimodaira H (2015) pvclust: Hierarchical Clustering with P-Values via Multiscale Bootstrap Resampling. Bioinformatics. 12: 1540-1542. https://doi.org/10.1093/bioinformatics/btl117
- Tusiime G (2003) Variation and detection of Fusarium solani f.sp. phaseoli and quantification of soil inoculum in common bean fields. Ph.D. thesis. Makerere University Kampala.
- Yokoyama K and Ogoshi A (1986) Studies on hyphal anastomosis of Rhizoctonia solani. IV. Observation of imperfect fusion by light and electron microscopy. Trans. Mycol. Soc. Jpn. 27: 399–413.
Keywords:
Download:
Full PDF- Share This