AtBTH1, a BTB domain-containing protein, influences sugar-responsive germination and seedling growth in Arabidopsis
Received: October 15, 2018
Accepted: October 22, 2018
Published: November 05, 2018
Genet.Mol.Res. 17(4): http://dx.doi.org/gmr16039932
DOI: http://dx.doi.org/10.4238/gmr16039932
Abstract
Sugars function as signal molecules that regulate gene expression, growth, and development in plants, animals, and yeasts. As a contribution to understanding the molecular mechanisms of sugar responses, we identified and characterized an Arabidopsis thaliana BTB domain-containing protein of the H subfamily 1 (designated AtBTH1). The relative expression level of AtBTH1 was enhanced by exogenous sugar treatment. To determine the function of AtBTH1 in sugar response, we identified two T-DNA insertion mutants, atbth1-1 and atbth1-2, which displayed an insensitive response to sugar-mediated inhibition of seed germination. The relatively early seedling growth of the atbth1-1 and atbth1-2 mutants was promoted by exogenous sugar treatment. Light-grown seedlings of the atbth1-1 and atbth1-2 mutants developed long roots when grown on medium containing a high sugar concentration. Dark-grown atbth1-1 and atbth1-2 seedlings showed sugar-insensitive hypocotyl elongation and development. These findings revealed that expression changes caused by mutation of AtBTH1 altered sugar-responsive seed germination and seedling growth. Taken together, the results suggest that AtBTH1 plays an important role in sugar responses in Arabidopsis.
Keywords
Arabidopsis thaliana; BTB protein; Sucrose; Glucose; T-DNA mutant; Seedling growth
Introduction
Plant growth is regulated by diverse developmental and environmental signals, and also is modulated by the physiological and metabolic condition of the plant. Sugars are essential substrates for carbon and energy metabolism, and function as signalling molecules to coordinate metabolic pathways (Rolland and Sheen, 2005). As primary products of photosynthesis, sugars influence the majority of, if not all, metabolic processes in plant cells by providing essential elements for organic compounds and energy for chemical reactions (Rolland et al., 2006; Smeekens and Hellmann, 2014).
The important role of sugars as signalling molecules and their regulatory effects on plant growth and development have been recognized in recent decades (Rolland et al., 2002; Rolland and Sheen, 2005). Sugar-responsive pathways typically involve genes that encode putative transcription factors and protein kinases, and genes required for protein synthesis and ubiquitin-mediated protein degradation (Bläsing et al., 2005; Gutierréz et al., 2008; Usadel et al., 2008). Plant G-box (CACGTG)-binding factors (GBFs) of the basic leucine zipper (bZIP) family of transcription factors, such as bZIP2/GBF5 and bZIP11/ATB1, in conjunction with Snf1-related kinases (SnRKs), such as KIN10/11, coordinate synergistic transcriptional networks promoted in response to sugar and energy deprivation as well as diverse stresses (Baena-González et al., 2007; Hanson et al., 2008). The Arabidopsis SnRK1 isoforms AKIN10 and AKIN11 are regulated by PRL1 (Pleiotropic Regulatory Locus-1), which is an evolutionarily conserved nuclear α-importin-binding WD protein (Nemeth et al., 1998; Bhalerao et al., 1999; Farrás et al., 2001). The prl1 mutation causes transcriptional derepression of many sucrose-regulated genes and results in diverse phenotypic responses, including arrested root elongation, altered leaf development, inhibition of cell elongation, and hypersensitivity to glucose, sucrose, and several hormones (Németh et al., 1998). However, the molecular mechanisms downstream of bZIP/SnRK that regulate the adapation responses remain unclear.
Broad-complex, Tramtrack, and Bric-a-brac (BTB) proteins have been described in plants that contain a highly conserved domain of approximately 115 amino acids (Stogios et al., 2005; Weber et al., 2005; Figueroa et al., 2005; Gingerich et al., 2005; Gingerich et al., 2007). The BTB domain, also termed the poxvirus and zinc finger (POZ) domain, is an evolutionarily conserved protein–protein interaction motif (Bardwell and Treisman, 1994; Cheng et al., 2014). The BTB domain is composed of six α-helices and three β-sheets that form a butterfly-shaped dimer with a tightly interwound peptide chain and has an extensive hydrophobic interface (Ahmad et al., 2003; Ahmad, 1998). The BTB proteins are perform crucial roles in plant development (Chevrier et al., 2014; Juranič et al., 2012; Robert et al., 2009; Xu et al., 2016), nitrogen use efficiency (Araus et al., 2016), seed germination (Kim et al., 2016), and responses to biotic and abiotic stresses (Boyl et al., 2009; Mandadi et al., 2009). In the Arabidopsis genome 80 genes that encode BTB domain-containing proteins have been identified and are classified into 10 subfamilies. Five BTB domain-containing genes classified in the H subfamily have been identified in Arabidopsis thaliana (AtBTH). The locus At2g13690 interacts with the WD protein PRL1, which has diverse pleiotropic effects on hormone and sugar responses and developmental processes in Arabidopsis (Németh et al., 199).However, At2g13690 is the only member of the H subfamily in Arabidopsis to have been studied intensively. Thus, further investigations are required to elucidate the functions of the other AtBTH genes.
In the present study, we identified and characterized AtBTH1 (At5g60050) of the H subfamily of BTB domain-containing proteins in Arabidopsis. The phylogenetic relationships and gene structure were compared among the Arabidopsis AtBTH genes. The expression profile of each AtBTH gene in response to exogenous sugar treatments was analyzed using quantitative real-time RT-PCR (qRT-PCR). Loss-of-function of AtBTH1 resulted in insensitivity to sugar during seed germination and seedling growth. Thus, AtBTH1 was indicated to be a key element in sugar response.
Material and Methods
Plant material and growth conditions
The wild-type, AtBTH1-1, and AtBTH1-2 Arabidopsis thaliana plants used in this study were of the Columbia-0 ecotype. Seeds of the AtBTH1-1 (SALK_051265C) and AtBTH1-2 (SALK_068122C) mutants were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.com). The T-DNA insertion was confirmed by PCR and sequencing. Seeds were surface-sterilized with 70% ethanol and 2% (v/v) household bleach, then washed at least five times with sterile water. The sterilized seeds were stratified at 4°C for 3 d in the dark, and germinated on Murashige and Skoog (MS) medium (Duchefa, Haarlem, The Netherlands) with pH 5.7 and supplemented with 2% sucrose, and either 1.2% agar or 0.25% phytagel.
Sequence alignment and phylogenetic analysis
Deduced amino acid sequences for the five AtBTH proteins were aligned with ClustalW implemented in MEGA 6 using the default settings (Tamura et al., 2013). Phylogeny reconstruction was conducted using the neighbour-joining (NJ) method (Saitou and Nei, 1987). Statistical support for the tree topology was assessed using the bootstrap method with 1000 replications (Felsenstein, 1985) and the nucleotide substitution evolutionary model was adopted.
RNA isolation, RT-PCR, and quantitative real-time RT-PCR
To analyze the expression patterns of AtBTH genes we used qRT-PCR. Total RNA was extracted from 10-day-old seedlings treated with 100 mM sucrose for 1, 6, 12, 24, and 36 h as well as from control (non-treated) plants. Total RNA was isolated using Trizol reagent in accordance with the manufacturer’s protocol (GibcoBRL, Cleveland, OH, USA). Total RNA (1 μg) of each sample extract was reverse transcribed using the Power cDNA Synthesis Kit (iNtRON Biotechnology Inc., Sungnam, Korea). The gene-specific primers used for RT-PCR and qRT-PCR are listed in Table 1. PCR amplification with each gene-specific primer was performed using 1 μl cDNA (Table 1). The resulting RT-PCR products were electrophoresed on a 1.0% (w/v) agarose gel after staining with ethidium bromide. Quantitative real-time RT-PCR was carried out on a Bio-Rad CFX real-time PCR detection system (Bio-Rad Laboratories Inc., Hercules, CA, USA) using iQ™ SYBR® Green Supermix (Bio-Rad). The reaction was performed under the following conditions: 95°C for 10 min, followed by 45 cycles of 95°C for 10 s and 60°C for 30 s. The relative expression levels were calculated using the relative 2−ΔΔCt method (Livak and Schmittgen, 2001). All real-time RT-PCR analyses were repeated with three biological and three technical replications.
| AGI No. | Target genes | Forward and reverse primers |
|---|---|---|
| For quantitative real-time RT-PCR | ||
| AT4g15900 | PRL1 | 5'- TCT GTT GCG TTT GAC CCG A |
| 5'- ACT CCA GTT GCT ACA TCC CAT | ||
| At5g60050 | AtBTH1 | 5'- TCG AAG CTT CTC ACT GGT GT |
| 5'- AGC ACA AGA GAC GTG AGA CA | ||
| At1g63850 | 5'- GAG AGC TTC GAA AGG GCT TG | |
| 5'- CTC CAC CAA ACT TCG AAC CC | ||
| At2g13690 | 5'- GCT CAC CGC AAA GCT CTA AA | |
| 5'- TGG TCT CGG TCT CTT GCA TT | ||
| At3g05675 | 5'- TCT GCA GAC CAA GCT GAG AT | |
| 5'- ACA TGC CGT TCT CCT CTT CA | ||
| At3g50780 | 5'- CCT GCT TGG ATT CGG TTC TG | |
| 5'- TTG GTT TGC CCA TGT GAC TG | ||
| At3g18780 | Actin2 | 5’- TCC CTC AGC ACA TTC CAG CA |
| 5’- GAT CCC ATT CAT AAA ACC CCA GC | ||
| For semi-quantitative RT-PCR | ||
| At5g60050 | AtBTH1 | 5'-GTTTCTGCAGCTATCTCGTTTGATGAA 5'-GGATCCTCAATAGAGTGTAATCTGCAGCCG |
| At3g18780 | Actin2 | 5′-ATGGCTGAGGCTGATGATATTCAA 5′-TTAGAAACATTTTCTGTGAACGATT |
Table 1. Primer sequences used for semi-quantitative and quantitative real-time RT-PCR.
Phenotypic analysis under sugar treatments
To compare germination frequencies, control and AtBTH1 mutant seeds were harvested on the same day. Seeds were surface-sterilized and sown on 0.25% (w/v) phytagel plates containing MS medium (pH 5.8) supplemented with 0%, 2%, 4%, 6%, 8%, or 10% sucrose. The plates were incubated at 25°C under a long-day photoperiod, and germination was scored based on radicle emergence 4 days after sowing. In each experiment, approximately 100 seeds were used, and triplicate experiments were carried out using independent seed lots.
For the seedling growth test, seeds were sown on MS medium supplemented with glucose or sucrose (0%, 2%, 4%, 6%, 8%, or 10%) and 0.7% Phyto agar (Duchefa), and incubated at 24°C under a 16 h light/8 h dark photoperiod for 7 day. To measure root length, seedlings were grown vertically on the specified medium under constant light. At least 20 seedlings were measured for each sample. For the dark condition, wild-type and mutant seeds were germinated on MS medium supplemented with sucrose (0%, 2%, 4%, 6%, 8%, or 10%), 1.2% agar or 0.25% phytagel, exposed to light for 18 h, and then grown vertically in complete darkness at 25°C for 7 day. Measurement of hypocotyl length followed a similar protocol to that used for root length measurement, except that the seedlings were grown in darkness.
Results
Sequence comparison and phylogenetic analysis of AtBTH proteins
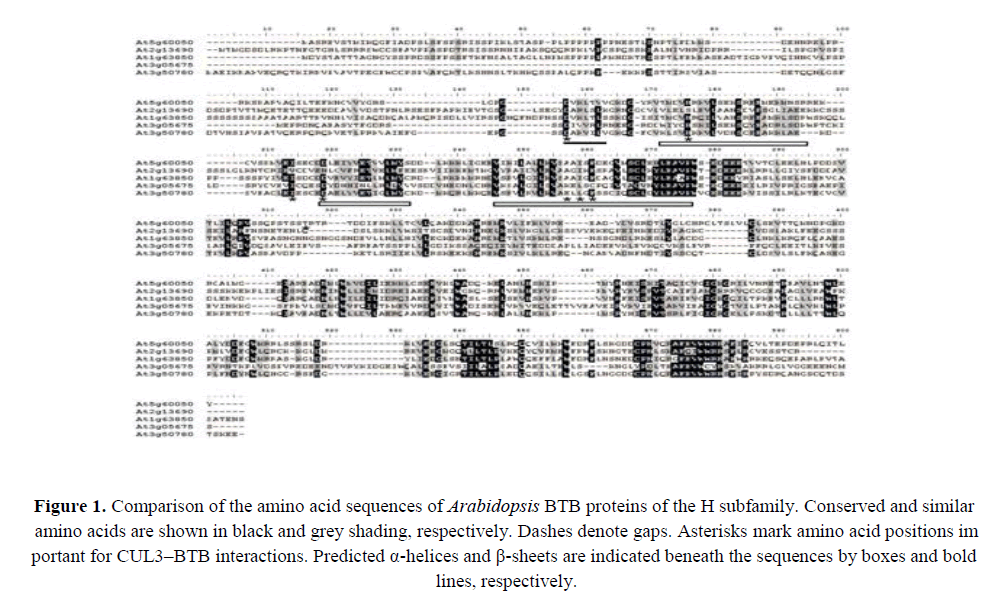
We generated an alignment of the deduced amino acid sequences of the BTB domain-containing H subfamily proteins in Arabidopsis (AtBTH proteins) (Figure 1). A consensus BTB motif that contained three conserved regions separated by short variable sequences was discernible (Figure 1). Residues in the BTB domains previously identified as important for binding CUL3 were located in the conserved regions, particularly Asp, His, Ile, Asp, Asp/Glu, Tyr, and Ile/Leu (Geyer et al., 2003, Xu et al., 2003). We noted that some of the predicted BTB proteins lacked one or more of these conserved residues (Figure 1). Specifically, the five members of the H subfamily all shared a ~300 amino acid region of 32%–63% similarity in the C-terminus of the BTB domain (Figure 1). This conserved region may be important for target recognition.
Figure 1. Comparison of the amino acid sequences of Arabidopsis BTB proteins of the H subfamily. Conserved and similar amino acids are shown in black and grey shading, respectively. Dashes denote gaps. Asterisks mark amino acid positions important for CUL3–BTB interactions. Predicted α-helices and β-sheets are indicated beneath the sequences by boxes and bold lines, respectively.
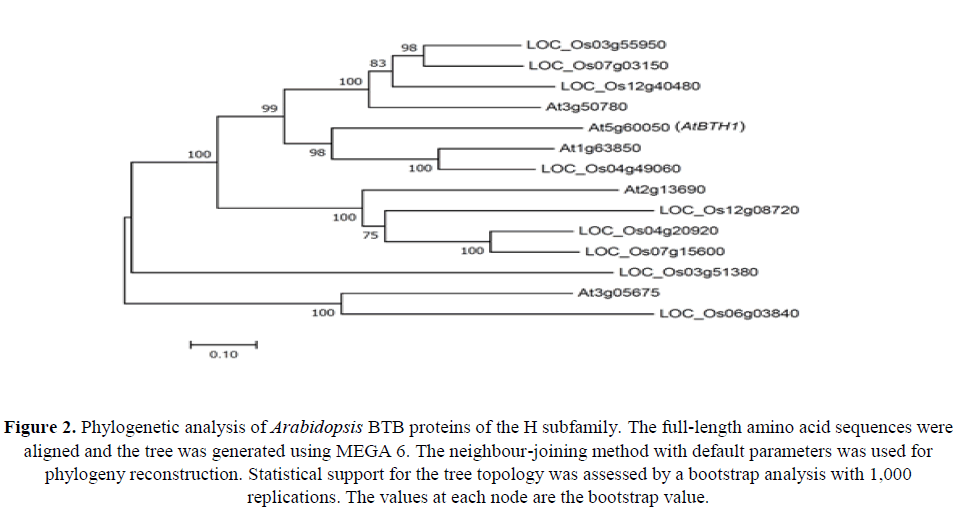
To determine phylogenetic relationships among BTB proteins of the H subfamily from Arabidopsis and the monocot rice (Oryza sativa), a phylogenetic analysis of deduced amino acid sequences was performed using the neighbour-joining method (Figure 2). The phylogenetic reconstruction showed that AtBTH1 (At5g60050) was grouped with At1g63850 and LOC_Os04g49060.
Figure 2. Phylogenetic analysis of Arabidopsis BTB proteins of the H subfamily. The full-length amino acid sequences were aligned and the tree was generated using MEGA 6. The neighbour-joining method with default parameters was used for phylogeny reconstruction. Statistical support for the tree topology was assessed by a bootstrap analysis with 1,000 replications. The values at each node are the bootstrap value.
Expression of AtBTH genes under sucrose treatment
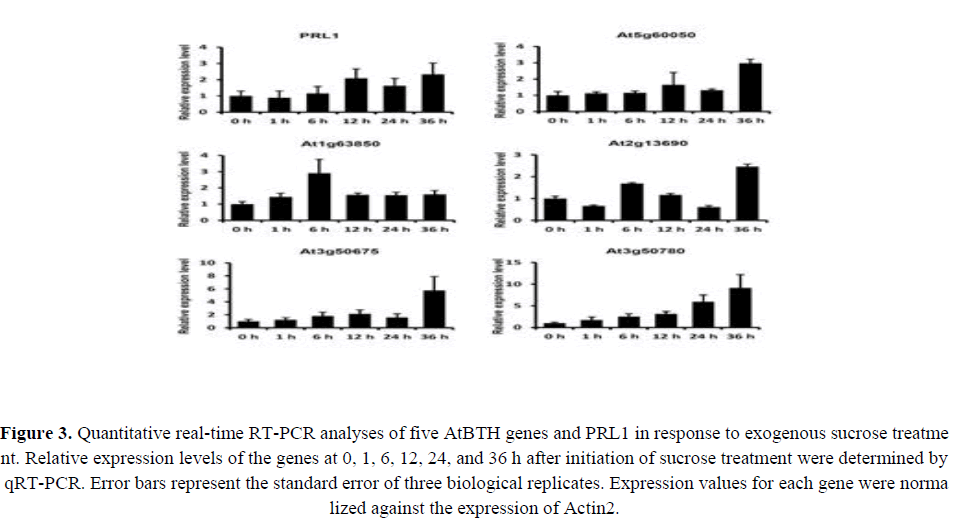
A previous study indicated the possible involvement of BTB genes in sugar responses (Mandadi et al., 2009). Gene expression patterns can provide important clues for gene function; therefore, we performed qRT-PCR to compare the expression patterns of the five AtBTH genes in response to exogenous sugar treatments. All AtBTH genes showed an increased expression level in the sugar treatments. This result supported the involvement of the AtBTH genes in responses to sugar. At1g63850 showed the highest expression level at 6 h, whereas the highest expression levels of At5g60050, At2g13690, At3g50675, and At3g50780 were observed at 36 h (Figure 3). Among these genes, the expression pattern of At5g60050 (AtBTH1) showed the greatest similarity to that of PRL1; the relative expression level increased after 12 h under sugar treatment (Figure 3). Thus, AtBTH1 was selected for additional investigation in this study.
Figure 3. Quantitative real-time RT-PCR analyses of five AtBTH genes and PRL1 in response to exogenous sucrose treatment. Relative expression levels of the genes at 0, 1, 6, 12, 24, and 36 h after initiation of sucrose treatment were determined by qRT-PCR. Error bars represent the standard error of three biological replicates. Expression values for each gene were normalized against the expression of Actin2.
Identification of T-DNA insertion mutants in AtBTH1
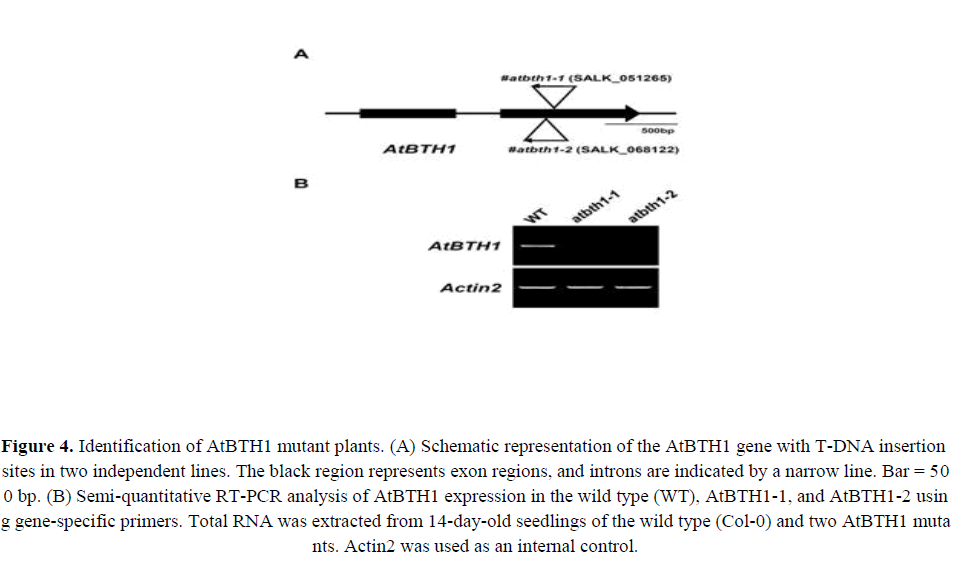
The AtBTH1 gene consists of 1500 bp between the start and stop codons. The gene is organized into two exons and one intron (Figure 4A). To determine the function of AtBTH1, we identified two independent T-DNA insertion mutants for AtBTH1. The AtBTH1-1 (SALK_051265) and AtBTH1-2 mutants (SALK_068122) both contain a T-DNA insertion in the second exon of AtBTH1 (Figure 4A). The two mutants had single T-DNA insertions (data not shown) that were predicted to disrupt AtBTH1 expression. This finding was confirmed by semi-quantitative RT-PCR (Figure 4B). The RT-PCR analysis showed that no PCR products were detected among the AtBTH1 mutant lines, which indicated that AtBTH1-1 and AtBTH1-2 likely represent loss-of-function alleles of AtBTH1 (Figure 4B). No obvious morphological differences were observed between wild-type and mutant plants.
Figure 4. Identification of AtBTH1 mutant plants. (A) Schematic representation of the AtBTH1 gene with T-DNA insertion sites in two independent lines. The black region represents exon regions, and introns are ndicated by a narrow line. Bar = 500 bp. (B) Semi-quantitative RT-PCR analysis of AtBTH1 expression in the wild type (WT), AtBTH1-1, and AtBTH1-2 using gene-specific primers. Total RNA was extracted from 14-day-old seedlings of the wild type (Col-0) and two AtBTH1 mutants. Actin2 was used as an internal control.
Insensitivity of AtBTH1 mutants to sugar stress
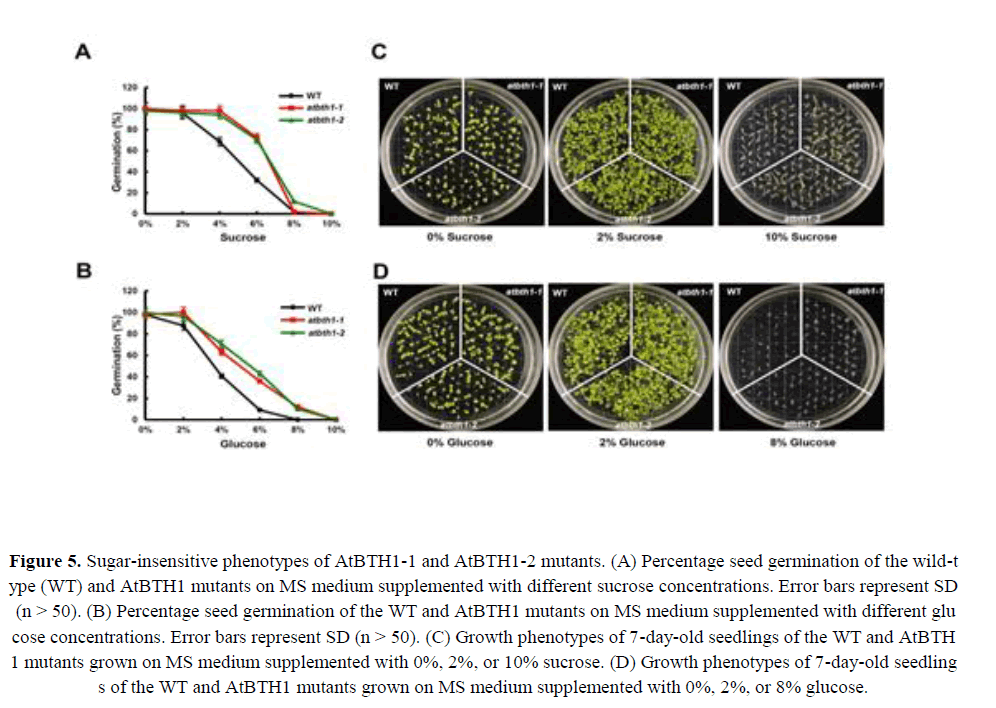
Sugars, in addition to their metabolic roles, act as signalling molecules and control key aspects of plant growth and development. Several mutants implicated in sucrose and glucose responses display decreased sensitivity to the inhibitory effects of high sugar concentrations on seed germination and seedling establishment (Dijkwel et al., 1996; Zhou et al., 1998; Huijser et al., 2000; Laby et al., 2000). Given that BT2 affects germination in the presence of sugar (Mandadi et al., 2009), we determined whether the BTB H subfamily gene, AtBTH1, also affected seed germination under exogenous sugar treatment. We germinated seeds from the AtBTH1-1 and AtBTH1-2 mutants and wild-type plants on medium supplemented with various concentrations of sucrose (0%, 2%, 4%, 6%, 8%, or 10%) or glucose (0%, 2%, 4%, 6%, 8%, or 10%) and the percentage cotyledon emergence was determined. All lines showed almost 100% germination on medium supplemented with 2% sucrose or 2% glucose. However, higher concentrations of sucrose and glucose were sufficient to inhibit germination of the AtBTH1-1 and AtBTH1-2 mutants (Figures 5A and 5B). The 8% and 10% concentrations of sucrose and glucose inhibited germination of all lines equally (Figures 5A and 5B).
Figure 5. Sugar-insensitive phenotypes of AtBTH1-1 and AtBTH1-2 mutants. (A) Percentage seed germination of the wild-type (WT) and AtBTH1 mutants on MS medium supplemented with different sucrose concentrations. Error bars represent SD (n > 50). (B) Percentage seed germination of the WT and AtBTH1 mutants on MS medium supplemented with different glucose concentrations. Error bars represent SD (n > 50). (C) Growth phenotypes of 7-day-old seedlings of the WT and AtBTH1 mutants grown on MS medium supplemented with 0%, 2%, or 10% sucrose. (D) Growth phenotypes of 7-day-old seedlings of the WT and AtBTH1 mutants grown on MS medium supplemented with 0%, 2%, or 8% glucose.
To check the early seedling growth of AtBTH1 mutants in response to sucrose and glucose, seeds of the wild type and the AtBTH1 mutants were sown on medium supplemented with various concentrations of sucrose and glucose. On medium that contained 2% sucrose or 2% glucose, the AtBTH1 mutants showed no difference in growth and development from those of the wild type. However, on media containing 10% sucrose or 8% glucose, the AtBTH1 mutants showed greater insensitivity than the wild type (Figure 5). The cotyledons and roots of the mutants were more uniformly developed than those of the wild type (Figures 5C and 5D). These results suggested that AtBTH1 controlled sugar signalling during germination and early seedling development.
Light-independent insensitive phenotype of AtBTH1 mutants in response to sucrose treatment
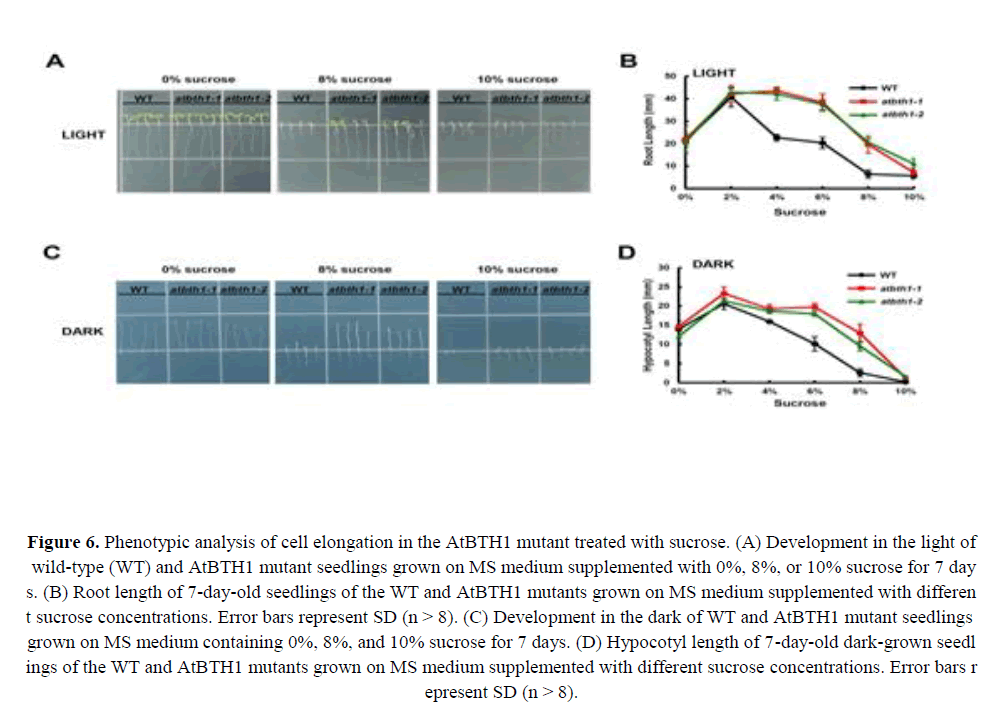
Dark-grown Arabidopsis seedlings develop leaf-like organs on vertical plates with exposure to extremely low concentrations of sugars, which suggests that the dark development phenotype of Arabidopsis seedlings is a sensitive indicator of the effects of sugars on plant growth and development (Roldán et al., 1999; Baier et al., 2004; Li et al., 2007; Zheng et al., 2015). To gain insight into how AtBTH1 influences sugar responses, we investigated the light or dark development phenotype of 7-day-old Col-0, AtBTH1-1, and AtBTH1-2 seedlings. With regard to the light condition, the mutants showed a dramatically insensitive phenotype of the roots and cotyledons compared with those of the wild type (Figures 6A and 6B).
Figure 6. Phenotypic analysis of cell elongation in the AtBTH1 mutant treated with sucrose. (A) Development in the light of wild-type (WT) and AtBTH1 mutant seedlings grown on MS medium supplemented with 0%, 8%, or 10% sucrose for 7 days. (B) Root length of 7-day-old seedlings of the WT and AtBTH1 mutants grown on MS medium supplemented with different sucrose concentrations. Error bars represent SD (n > 8). (C) Development in the dark of WT and AtBTH1 mutant seedlings grown on MS medium containing 0%, 8%, and 10% sucrose for 7 days. (D) Hypocotyl length of 7-day-old dark-grown seedlings of the WT and AtBTH1 mutants grown on MS medium supplemented with different sucrose concentrations. Error bars represent SD (n > 8).
In dark-grown AtBTH1 seedlings, hypocotyl length showed no difference from that of wild-type plants in response to 0% and 4% sucrose treatment, and elongation was progressively elevated with increase in sucrose concentration between 6% and 10% (Figure 6D). Hypocotyl elongation in AtBTH1 seedlings was similar to that of wild-type seedlings at no or low sucrose concentrations, but at higher sucrose concentrations hypocotyl length was progressively longer than that of the wild type (Figures 6C and 6D). These results indicated that the mutations in AtBTH1 might affect insensitivity to sucrose and light independently.
Discussion
Sugars modulate multiple responses in plant growth, development, gene expression, and metabolism (Rolland et al., 2006). BTB proteins are central components of an interconnected signalling network that detects and responds to multiple inputs, such as nutrients, stresses, and hormones (Mandadi et al., 2009). However, the function of AtBTH genes has not been elucidated.
The present results showed that the expression level of the five AtBTH genes was affected by exogenously applied sugars, which was confirmed in the qRT-PCR analysis. All of the studied genes showed a similar expression pattern to that of PRL1 (Figure 3). PRL1 is a component of the SnRK1 pathway that encodes an E3 ubiquitin ligase and interacts with At2g13690 of the BTB H subfamily (Nemeth et al., 1998). This suggests that AtBTH genes play roles in the regulation of sugar responses, at least at the transcriptional level. In addition, AtBTH genes may influence the sugar response through the SnRK1 pathway. Among these genes, At5g60050 (AtBTH1) showed the greatest similarity in expression pattern to that of PRL1, with the expression level increasing after 12 h under sugar treatment (Figure 3). Therefore, AtBTH1 was selected for further investigation in this study.
The mRNA expression level of four AtBTH genes, but not that of At3g50780, was decreased at 24 h compared with that at 12 h (Figure 3). Previous studies have demonstrated that BT2 is controlled by light (Mandadi et al., 2009). Consistent with this finding, the peaks in AtBTH transcript levels occurred during the light period at 6, 12, or 36 h (Figure 3). These results suggest that light may also modulate AtBTH transcription.
To further investigate AtBTH1 gene function in response to sugar, we analyzed seed germination in the presence of exogenous glucose and sucrose, and observed that loss of AtBTH1 function resulted in decreased sensitivity to high concentrations of sugars. After identifying a role for AtBTH1 in modulation of sugar responses during seed germination, we investigated whether seedling growth also was modulated by AtBTH1. Thus, our findings define an important genetic and molecular mechanism mediated by AtBTH1 to regulate sugar-responsive germination in Arabidopsis.
During seedling growth and development, high concentrations of sugar induce developmental arrest in the light (Arenas-Huertero et al., 2000). Light-grown AtBTH1 mutant seedlings displayed typical physiological changes caused by exogenously applied sugars or decreased sugar sensitivity (Figure 5). Development in the dark is a sensitive indicator of the effects of sugars on growth and development in Arabidopsis (Baier et al., 2004; Li et al., 2007). Hypocotyl elongation of dark-grown Arabidopsis seedlings is progressively inhibited by high concentrations of sugar (Li et al., 2007). In the present study, we observed that dark-grown AtBTH1-1 and AtBTH1-2 mutants exhibited increased hypocotyl length in response to exogenous sucrose treatment (Figure 6). Mutations in PRL1 (At4g15900) lead to transcriptional derepression of glucose-responsive genes and increase the sensitivity of plants to growth hormones, including cytokinin, ethylene, abscisic acid, and auxin (Németh et al., 1998). Given that the Arabidopsis prl1 mutant shows hypersensitivity to sugars in light and dark conditions (Németh et al., 1998; Li et al., 2007). The pattern of hypocotyl elongation of AtBTH1 mutants in the dark (Figure 6C) was essentially different to that of the prl1 mutant. This observation suggested that the sugar-responsive pathways that control dark development and hypocotyl elongation in wild-type seedlings show an antagonistic function with that of PRL1.
Conclusion
The present results demonstrated that AtBTH1 plays a crucial role in regulating sugar-responsive growth and development under both light and dark conditions. These findings will help to provide an improved mechanistic understanding of the role of AtBTH1 in sugar-induced responses. In summary, we present evidence that AtBTH1 encodes a functionally unknown protein involved in sugar-related responses in Arabidopsis. In the future, genome-wide transcriptomic and proteomic analyses combined with studies of protein–protein interaction in vitro and in vivo will provide additional information for functional analysis of AtBTH1 in the sugar response.
Acknowledgment
This research was supported by the National Institute of Ecology.
Conflicts of interest
The authors declare no conflict of interest.
About the Authors
Corresponding Author
H.C. Park
Division of Ecological Conservation, Bureau of Ecological Research, National Institute of Ecology, Seocheon, Republic of Korea
- Email:
- jiha@gntech.ac.kr
References
- Ahmad KF, Engel CK, Privé GG (1998) Crystal structure of the BTB domain from PLZF. Proc Natl Acad SciUSA. 95: 12123-12128. https://doi.org/10.1073/pnas.95.21.12123
- Ahmad KF, Melnick A, Lax S, Bouchard D, et al. (2003) Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol Cell 12: 1551-1564. https://doi.org/10.1016/s1097-2765(03)00454-4
- Araus V, Vidal EA, Puelma T, Alamos S, et al., (2016) Members of BTB gene family of scaffold proteins suppress nitrate uptake and nitrogen use efficiency. Plant Physiol 171: 1523-1532. https://doi.org/10.1104/pp.15.01731
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, et al. (2000) Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev 14: 2085-2096.
- Baena-González E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938-942. https://doi.org/10.1038/nature06069
- Baier M, Hemmann G, Holman R, Corke F, et al. (2004). Characterization of mutants in Arabidopsis showing increased sugar-specific gene expression, growth, and developmental responses. Plant Physiol 134: 81-91. https://doi.org/10.1104/pp.103.031674
- Bardwell VJ and Treisman R (1994) The POZ domain: a conserved protein-protein interaction motif. Genes Dv 8: 1664-1677. https://doi.org/10.1101/gad.8.14.1664
- Bhalerao RP, Salchert K, Bakó L, Okresz L, et al. (1999) Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1- like protein kinases. Proc Natl Acad Sci USA 96: 5322-5327. https://doi.org/10.1073/pnas.96.9.5322
- Bläsing OE, Gibon Y, Günther M, Höhne M, et al. (2005) Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17: 3257-3281. https://doi.org/10.1105/tpc.105.035261
- Boyle P, Le Su E, Rochon A, Shearer HL, et al. (2009) The BTB/POZ domain of the Arabidopsis disease resistance protein NPR1 interacts with the repression domain of TGA2 to negate its function. Plant Cell 21: 3700-3713. https://doi.org/10.1105/tpc.109.069971
- Cheng D, Qian W, Meng M, Wang Y, et al. (2014) Identification and expression profiling of the BTB domain-containing protein gene family in the silkworm, bombyx mori. Int J Genom 2014: 1-14. https://doi.org/10.1155/2014/865065
- Chevrier S, Emslie D, Shi W, Kratina T, et al. (2014) The BTB-ZF transcription factor Zbtb20 is driven by Irf4 to promote plasma cell differentiation and longevity. J Exp Med 211: 827-840. https://doi.org/10.1084/jem.20131831
- Dijkwel PP, Kock PAM, Bezemer R, Weisbeek PJ, et al. (1996) Sucrose represses the developmentally controlled transient activation of the plastocyanin gene in Arabidopsis thaliana seedlings. Plant Physiol. 110: 455-463. https://doi.org/10.1104/pp.110.2.455
- Farrás R, Ferrando A, Jásik J, Kleinow T, et al. (2001) SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. Embo J 20: 2742-2756. https://doi.org/10.1093/emboj/20.11.2742
- Felsenstein J (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783-791. https://doi.org/10.2307/2408678
- Figueroa P, Gusmaroli G, Serino G, Habashi J, et al. (2005) Arabidopsis has two redundant Cullin3 proteins that are essential for embryo development and that interact with RBX1 and BTB proteins to form multisubunit E3 ubiquitin ligase complexes in vivo. Plant Cell 17: 1180-1195. https://doi.org/10.1105/tpc.105.031989
- Gingerich DJ, Gagne JM, Salter DW, Hellmann H, et al. (2005) Cullins 3a and 3b assemble with members of the broad complex ⁄tramtrack⁄bric-a-brac (BTB) protein family to form essential ubiquitin-protein ligases (E3s) in Arabidopsis. J Biol Chem 280: 18810-18821. https://doi.org/10.1074/jbc.m413247200
- Gingerich DJ, Hanada K, Shiu SH, Vierstra RD (2007) Large-scale, lineage-specific expansion of a brica-brac ⁄tramtrack ⁄broad complex ubiquitin-ligase gene family in rice. Plant Cell 19: 2329-2348. https://doi.org/10.1105/tpc.107.051300
- Geyer R, Wee S, Anderson S, Yates J, et al. (2003) BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol Cell 12:783-790. https://doi.org/10.1016/s1097-2765(03)00341-1
- Gutierréz RA, Stokes TL, Thum K, Xu X, et al. (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci 105:4939-4944. https://doi.org/10.1073/pnas.0800211105
- Hanson J, Hanssen M, Wiese A, Hendriks MM, et al. (2008) The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of Asparagine Synthetase-1 and Proline Dehydrogenase-2. Plant J 53:935-949. https://doi.org/10.1111/j.1365-313x.2007.03385.x
- Huijser C, Kortstee A, Pego J, Weisbeek P, et al. (2000) The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: involvement of abscisic acid in sugar responses. Plant J 23:577-585. https://doi.org/10.1046/j.1365-313x.2000.00822.x
- Juranič M, Srilunchang KO, Krohn NG, Leljak-Levanic D, et al. (2012) Germline-specific MATH-BTB substrate adaptor MAB1 regulates spindle length and nuclei identity in maize. Plant Cell 24: 4974-4991. https://doi.org/10.1105/tpc.112.107169
- Kim H, Kim SH, Seo DH, Chung S, et al. (2016) ABA-HYPERSENSITIVE BTB/POZ PROTEIN 1 functions as a negative regulator in ABA-mediated inhibition of germination in Arabidopsis. Plant Mol Biol 90: 303-315. https://doi.org/10.1007/s11103-015-0418-7
- Laby RJ, Kincaid MS, Kim D, Gibson SI (2000) The Arabidopsis sugar insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J 23: 587-596. https://doi.org/10.1046/j.1365-313x.2000.00833.x
- Li Y, Smith C, Corke F, Zheng L, et al. (2007) Signaling from an altered cell wall to the nucleus mediates sugar responsive growth and development in Arabidopsis thaliana. Plant Cell 19: 2500-2515. https://doi.org/10.1105/tpc.106.049965
- Mandadi KK, Misra A, Ren S, McKnight TD (2009) BT2, a BTB protein, mediates multiple responses to nutrients, stresses, and hormones in Arabidopsis. Plant Physiol 150: 1930-1939. https://doi.org/10.1104/pp.109.139220
- Németh K, Salchert K, Putnoky P, Bhalerao R, et al. (1998) Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev 12: 3059-3073. https://doi.org/10.1101/gad.12.19.3059
- Rolland F and Sheen J (2005) Sugar sensing and signaling networks in plants. Biochem Soc Trans 33: 269-271. https://doi.org/10.1042/bst0330269
- Robert HS, Quint A, Brand D, Vivian-Smith A, et al. (2009) BTB and TAZ domain scaffold proteins perform a crucial function in Arabidopsis development. Plant J 58: 109-121. https://doi.org/10.1111/j.1365-313x.2008.03764.x
- Roldán M, Gómez-Mena C, Ruiz-García L, Salinas J, et al. (1999) Sucrose availability on the aerial part of the plant promotes morphogenesis and flowering in the dark. Plant J 20: 581-590. https://doi.org/10.1046/j.1365-313x.1999.00632.x
- Rolland F, Moore B, Sheen J (2002) Sugar sensing and signaling in plants. Plant Cell 14: S185-S205. https://doi.org/10.1105/tpc.010455
- Rolland F, Baena-Gonzalez E, Sheen J (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu. Rev. Plant Biol 57: 675-709. https://doi.org/10.1146/annurev.arplant.57.032905.105441
- Saitou N and Nei M (1987) The neighbour-joining method: a new method for reconstructing phylogenetic trees. Molec. Biol Evol 4: 406-425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
- Smeekens S and Hellmann HA (2014) Sugar sensing and signaling in plants. Front Plant Sci 5: 113. https://doi.org/10.3389/fpls.2014.00113
- Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, et al. (2005) Sequence and structural analysis of BTB domain proteins. Genome Biol 6: R82. https://doi.org/10.1186/gb-2005-6-10-r82
- Tamura K, Stecher G, Peterson D, Filipski A, et al. (2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30: 2725-2729. https://doi.org/10.1093/molbev/mst197
- Usadel B, Bläsing OE, Gibon Y, Retzlaff K, et al. (2008) Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol. 146: 1834-1861. https://doi.org/10.1104/pp.107.115592
- Weber H, Bernhardt A, Dieterle M, Hano P, et al. (2005) Arabidopsis AtCUL3a and AtCUL3b form complexes with members of the BTB⁄POZ-MATH protein family. Plant Physiol 137: 83-93. https://doi.org/10.1104/pp.104.052654
- Xu C, Park SJ, Van Eck J, Lippman ZB (2016) Control of inflorescence architecture in tomato by BTB/POZ transcriptional regulators. Genes Dev 30: 2048-2061. https://doi.org/10.1101/gad.288415.116
- Xu L, Wei Y, Reboul J, Vaglio P, et al. (2003) BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425: 316-321. https://doi.org/10.1038/nature01985
- Zheng L, Shang L, Chen X, Zhang L, et al. (2015) TANG1, encoding a Symplekin_C domain-contained protein, influences sugar responses in Arabidopsis. Plant Physiol 168: 1000-1012. https://doi.org/10.1104/pp.15.00288
- Zhou L, Jang JC, Jones TL, Sheen J (1998) Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc Natl Acad Sci 95: 10294-10299. https://doi.org/10.1073/pnas.95.17.10294
Keywords:
Download:
Full PDF- Share This