Characterization of aapP and nopP genes related to the biological nitrogen fixation efficiency with soybean in contrasting strains of Bradyrhizobium japonicum
Received: November 14, 2017
Accepted: December 08, 2017
Published: January 12, 2018
Genet.Mol.Res. 17(1): gmr16039867
DOI: 10.4238/gmr16039867
Abstract
Soybean (Glycine max) is considered one of the most economically important agricultural crops in Brazil. Due to the high protein content of its grains, this crop demands a greater amount of nitrogen (N) for production. In general, for soybean, nitrogen fertilization costs through chemical fertilizers are considered high. However, the biological nitrogen fixation (BNF) through the use of biological inoculants, such as the strains of Bradyrhizobium genus, a rhyzobacterium, represents a viable alternative for soybean crops in Brazil and dismisses nitrogen fertilization. Because of the need to constantly increase soybean crop productivity, it is necessary to find more efficient Bradyrhizobium genus strains for BNF. Genes related to soybean BNF have been utilized as molecular markers in selection programs that aim to obtain more efficient bacterial strains in this process. Therefore, this study aimed to characterize the gens related to BNF efficiency (nopP, aapP, ilvD and bll3106) by PCR-RFLP in Bradyrhizobium japonicum strains. The nopP and aapP genes were sequenced and submitted to similarity analysis with genes deposited on the Genbank (NCBI). The results showed the presence of highly conservative sequences in nopP and aapP genes between the analyzed B. japonicum strains. The occurrence of these preserved nucleotide sequences can be related to the adaptive function that they can have during the evolutionary process of plant growth promoting rhizobacteria.
Introduction
Soybean is considered one of the most important agricultural crops in economy in Brazil. Soybean grains have high protein content and are one of the main sources of vegetal oil in the world (Hungria et al., 2007). Soybean has symbiosis with bacterial species found in the soil such as diazothrophic bacteria which are called rhyzobacteria. These bacteria make nitrogen available to plant through biological nitrogen fixation (BNF) of the soybean (Chueire et al., 2003). The utilization of these biological inoculants, which has already been adopted by some soybean producers, has increased crop productivity besides reducing environmental damages caused by chemical fertilization such as the contamination of the soil, river springs and groundwater (Hungria et al., 2007).
Rhyzobacteria associate themselves with soybean roots in symbiosis so that the plant supplies energy for the bacterium growth which in exchange provides nitrogen to the plant (Hungria et al., 2007). The main soybean symbionts are the bacteria from the Bradyrhizobium genus, and in Brazil the most representative and utilized species for BNF in commercial inoculants in soybean crop are Bradyrhizobium japonicum and Bradyrhizobium elkanii (Hungria et al., 2007).
These diazothrophic bacteria have an enzymatic complex called nitrogenases, capable of converting atmospheric nitrogen into ammonia, which can be incorporated by the plant in different forms of organic nitrogen. When in symbiosis with the host plant, bacteria promote the formation of nodules in their roots where BNF occurs. The nodulation factors (NOD factors) involve specific gene expressions called nodulation genes (NOD genes) that occur in the host plant as well as in the bacterium (Bortolan et al., 2009). The NOD gene transcription by rhyzobacteria controlled by the activation of different proteins that are then activated by flavonoids secreted by the plant, resulting in the production of NOD factors by the bacterium (Souza, 2006).
The soybean inoculation with Bradyrhizobium strains may result in more than 6.6 billion dollars per harvest (Hungria et al., 2007), but the constant development of agricultural practices, followed by frequent releases of soybean cultivars that are more productive and better adapted to different environmental conditions as well as the presence of strain populations in previously inoculated soils, induce the continuous search for more efficient Bradyrhizobium genotypes in BNF process (Santos et al., 2006; Hungria et al., 2007).
In order to conduct this search, it is necessary to know and understand the existing genes and proteins in different Bradyrhizobium genotypes related to the efficiency in the BNF process, which can be utilized as genotypical markers when selecting or obtaining more promising Bradyrhizobium commercial inoculants (Barcellos et al., 2009).
Barcellos et al. (2009) identified a series of B. japonicum genes that are possibly related to the BNF efficiency with soybean by representative difference analysis (RDA) such as the ones called nopP and aapP. The nopP gene codifies a protein of the secretion system III, nopP (nodulation outer protein), possibly related to the infection process of the host plant and, therefore, to BNF efficiency. The aapP gene codifies one of the proteins that compose an aminoacid transport system located in the plasmatic membrane, initially identified in Rhizobium leguminosarum, and called Aap L-amino acid ABC transporter, identified as permeases and which transports a great variety L-aminoacids. According to Barcellos et al. (2009), this aminoacid transport system can also be related to the efficiency process of soybean BNF with B. japonicum. After the identification of these genes, their nucleotide characterization is necessary. Thus, this study aimed to characterize nopP , aapP, ilvD and bll3106 genes found in contrasting B. japonicum strains for BNF efficiency with soybean for potential utilization as genotypical markers in the selection of more promising genotype with possibility of use as commercial inoculants.
MATERIAL AND METHODS
Biological material
The utilized Bradyrhizobium japonicum strains were from the crop collection of the the Brazilian Microbiological Resource Center (BMRC) (http://www.bmrc.lncc.br/), EmbrapaSoja (Londrina/Paraná/Brazil). Fourteen strains of B. japonicum were utilized: CPAC7, S204, S340, S370, S372, S406, S452, S468, S478, S490, S516, SEMIA586, USDA110 and USDA6. Regarding the efficiency with soybean, S370 and S516 were contrasting strains (Hungria et al., 1998).
Cultivation media and growth conditions
The strains were cultivated in YMA (yeast extract, mannitol and agar) or YM (yeast extract, mannitol, liquid medium) cultivation medium (VINCENT, 1970). The cultures were maintained in YM (containing 30% glycerol) at -70°C for long-term storage and at 4°C for use in cultivations (Ribeiro et al., 2009).
DNA extraction
DNA extraction was done according to Barcellos et al. (2009). The bacteria were cultivated in 100 mL of YM medium at 28°C until reaching exponential phase (109 mL-1 cells). The culture was centrifuged at 3000 g for 10 min and the pellet was re-suspended in 50 mL of TES (50 mM Tris-HCl, pH 8.0, 20 mM EDTA, pH 8.0, 200 mM NaCl). The centrifugation was repeated, and the pellet was re-suspended in 10 mL of TES. A 100 μL aliquot of lysozyme (Sigma®) (200 μg mL-1) and 30 μL of RNAse (1 mg mL-1) was added and the mixture was incubated at 30°C for 4 h. After this period, 10 μL of proteinase-K (20 μg μL-1) was added and the mixture was incubated for 12 h at 37°C. An equivalent volume (10 mL) of balanced phenol (pH 8.0) was added and the tube was manually inverted for 30 min. The mixture was centrifuged for 15 min (13000 g) at room temperature (22°C). The supernatant was transferred to another tube and, a volume of phenol, equivalent to the transferred supernatant, was added to it and homogenized for 20 min. Next, 5 mL of chloroform was added, and the mixture was homogenized by manual inversion for 10 min and centrifuged at 13000 g for 15 min at room temperature (22°C). The superior phase of the mixture was transferred to a new tube and a volume equivalent of isopropanol was added and homogenized for 5 min, followed by centrifugation at 3000 g for 15 min. Isopropanol was removed and DNA was dried at room temperature (22°C) for 20 min. The DNA was re-suspended in 50 μL of TE and incubated for 12 h at 4°C and posteriorly stored at –20°C. Finally, the extracted DNA was quantified by electrophoresis in 1% agarose gel, utilizing as concentration standard lambda phage DNA (Invitrogen®) in different concentrations. The quantified DNA was diluted to 5 ng μL-1 concentration for utilization.
Synthesis of oligonucleotides (primers)
For PCR amplification of selected genes, primers based on identified genes with greater similarity with the genome of B. japonicum USDA 110 strain whose genome is completely sequenced (Kaneko et al., 2002), as shown in Table 1. Fast PCR 6.0 program (Kalendar et al., 2009) utilizing recommended parameters by the manufacturer was utilized.
| Gene | Annealing temperature (oC) | Forward sequence | Reverse sequence | Size of expected sequence (bp) |
|---|---|---|---|---|
| aapP | 59 | 5’ – atgtccgaccccatcgtcaag – 3’ | 5’ – accgcagaatctggctcagga – 3’ | 736 |
| nopP | 59 | 5’ – TGAGATCGACCAGCTTTCCT – 3’ | 5’ – TGACGGATCATCTTCGTCAG – 3’ | 588 |
| ilvD | 58 | 5’ – ccaacatcaagcagaggctgc – 3’ | 5’ – cgcatagcactgtttctcgtgc – 3’ | 1703 |
| bll3106 | 60 | 5’ – cggattgcgaatgaccacccct – 3’ | 5’ – gttgttctccgcaaagctgct – 3’ | 3040 |
Source: Authors.
Table 1: Characteristics for PCR amplifications of nopP, aapP, ilvD and bll3106 genes and the size of the fragment generated in Bradyrhizobium japonicum USDA110 strain.
PCR amplifications
PCR amplifications were performed following the parameters described by Ribeiro et al. (2009). For each reaction, a volume of 50 μL: dNTPs (300 mM of each), PCR buffer (Tris base 20 mM, pH 8.4 and 50 mM KCl), primers (15 pmol of each); Taq DNA polymerase (1.0 U) and DNA (20 ng). The amplification cycles were done according to Ribeiro et. al. (2009), with modifications in the annealing temperature of primers, and the content of G/C of each utilized pair of primers, as shown in Table 1. Initially, a denaturation cycle occurred at 95°C for 2 min, followed by 35 denaturation cycles at 94°C for 45 s, annealing at 58–60°C (depending on the primer pair) for 45 s and extension at 72°C for 2 min, and to end up the amplification of a final extension cycle at 72°C for 5 min. All the PCR reactions were performed in a Master Cycler Gradient thermocycler (Eppendorf®). The amplification analyses of PCR products were done after electrophoresis in 1% agarose gel at 70 V for 3 h utilizing DNA Ladder 1 kb plus (Life Technologies®) as a molecular mass marker. The gels were bleached in ultrapure water for 15 min and photographed in a UV transluminator.
Restriction analysis (PCR-RFLP) of amplified genes
The nucleotide sequences of analyzed genes obtained from the genome of B. japonicum USDA110 strains (NCBI - http://www.ncbi.nlm.nih.gov/nuccore/27375111) were submitted to in silico analyses. Information on the enzymes utilized in PCR-RFLP analysis and the identification of cut loci of the number and size of generated fragments were obtained from Restriction Mapper program (http://www.restrictionmapper.org/).
The PCR amplification products were obtained by adding reactions of 1/10 of the volume of 3 M sodium acetate, 3 volumes of cold ethanol, and kept in ice for 5 min. After this period, the samples were centrifuged at 15000 g for 5 min. The ethanol was discarded and the precipitated DNA was re-suspended in 30 μL of ultrapure water, and stored at -20°C for posterior cut with restriction enzymes. The obtained PRC products were individually digested with three different restriction enzymes:
MSEI, AluI and SalI (Table 2). The digestion conditions followed the manufacturer’s specifications (Invitrogen®). The resulting products of restriction (RFLP) were analyzed after horizontal electrophoresis in 3% agarose gel at 65 V for 4 h.
| Strains | nopP | aapP | ilvD | bll3106 |
|---|---|---|---|---|
| S204 | + | + | - | - |
| S340 | + | + | - | - |
| S370 | + | + | - | - |
| S372 | + | + | - | - |
| S406 | + | + | - | - |
| S452 | + | + | - | - |
| S468 | + | + | - | - |
| S478 | + | + | - | - |
| S490 | + | + | - | - |
| S516 | + | + | - | - |
| USDA 110 | + | + | - | - |
| USDA 6 | + | + | - | - |
+ Gene amplification; - Negative for gene amplification. Source: Authors
Table 2: Amplified PCR sequences of nopP, aapP, ilvD and bll3106 in Bradyrhizobium japonicum strains.
Results and Discussion
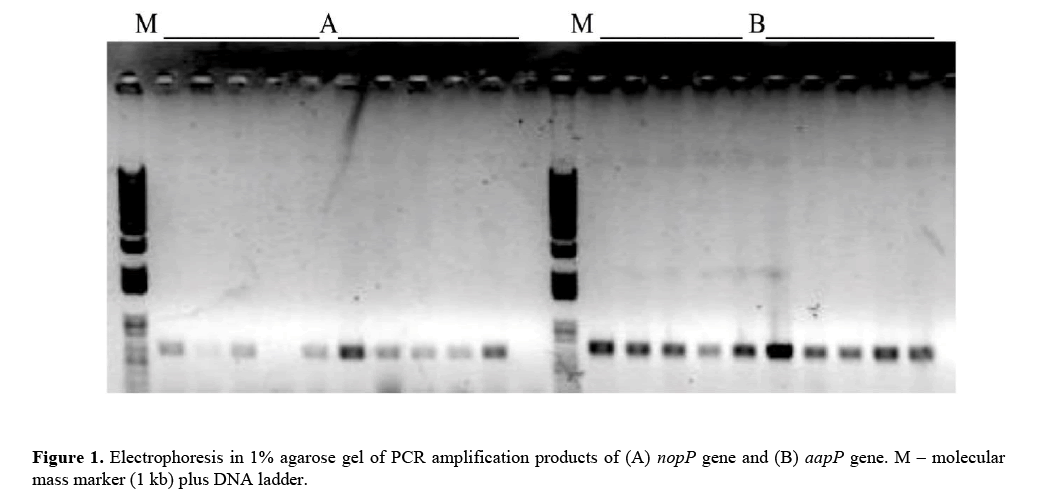
The synthesized primers based on the identified genes with greater similarity with the genome of B. japonicum USDA 110 strains are shown in Table 1. B. japonicum nopP , aapP, ilvD and bll3106 genes, described by Barcellos et al. (2009), were amplified by PCR technique (Table 2). Products of this amplification were obtained only for nopP and aapP genes for most evaluated strains, even after several attempts with modifications at annealing temperatures of primers and amount of utilized DNA (Figure 1). No amplification product was obtained for ilvD and bll3106 genes in the analyzed strains (Table 2), suggesting that these gene sequences be distinct from the ones found in the analyzed strains.
Positive amplifications were obtained for nopP and aapP genes although the amount of amplicons had been higher in the first one. Amplicons for nopP gene as well as for aapP gene presented the expected size (bp) for all evaluated strains, as previously identified in the genome of the strain utilized to build these primers, USDA 110 (Table 1). Size differences of the amplicons were not observed between the analyzed strains (Figure 1).
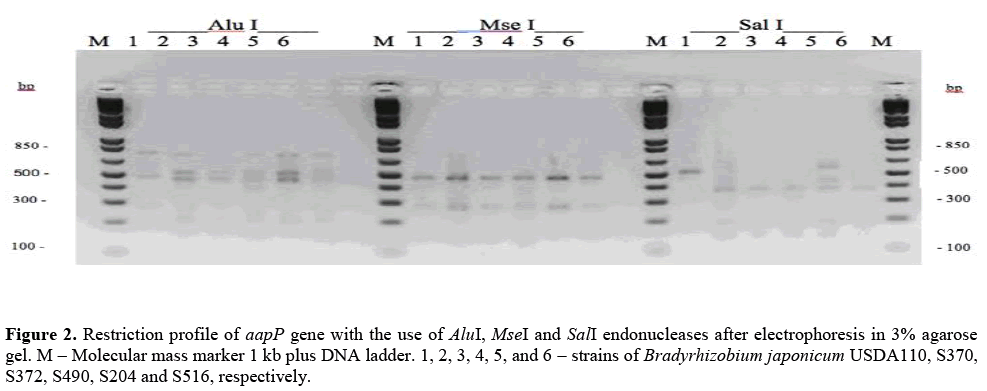
A low amount of amplicons in nopP gene was observed, even after the reamplification procedure has been done. So, due to the low amount of amplicons in nopP gene, only aapP gene was utilized in PCR-RFLP analyses. Three different endonucleases were applied (Table 3) and a low polymorphism level was verified among the analyzed strains (Figure 2), showing a high conservation level in the nucleotide sequences of this gene.
| Gene | Restriction enzyme | Locus cut number | Generated fragments (bp) |
|---|---|---|---|
| aapP | Mse I | 1 | 467, 269 |
| Alu I | 1 | 470, 266 | |
| Sal I | 2 | 510, 190, 36 |
Source: Authors.
Table 3: Restriction enzymes utilized in PCR-RFLP analyses of aapP gene, locus cut number and generated fragment sizes.
USDA110 strain presented a restriction standard of aapP gene with AluI and SalI endonucleases, differently from the other strains of B. japonicum (Figure 2), showing that the aapP gene of USDA110 strain presents differences in the nucleotide sequences when compared to the other utilized strains. This fact can explain the difficulty of PCR amplification of this gene in B. japonicum strains, once the primers utilized in the amplifications were based on the nucleotide sequences of the gene that was originally identified in USDA110 strain.
The restriction profile of aapP gene with SalI enzyme of S204 strain presented two additional bands of approximately 450 bp and 600 pb, besides the common bands of the other strains, indicating that this strain has two copies of aapP gene.
According to Delamuta et al. (2013), USDA110 strain belongs to Bradyrhizobium diazoefficiens (NCBI-http://www.ncbi.nlm.nih.gov/nuccore/27375111) and not to B. japonicum as originally identified. Our results corroborate the studies of Delamuta et al. (2013) once the observed restriction standards for the USDA110 strain are distinct from those observed for B. japonicum strains identified here.
Analyzing the similarities among the nucleotide sequences of aapP gene (obtained from GenBank–NCBI), through BLAST program (http://blast.ncbi.nlm.nih.gov) among several strains of different rhyzobacteria (genus Bradyrhizobium, Rhizobium, Sinorhizobium, Mesorhizobium, Azorhizobium, among them), a similarity level over 95% was observed among all analyzed species (unreported data). This result shows a high preservation level of this gene during the evolutionary process in these rhyzobacterium species, probably due to an adaptive factor of this gene during the evolutionary process.
The same similarity analysis by BLAST program, in silico analysis, was applied to nucleotide sequences of nopP genes among the two strains of B. japonicum with completely sequenced genome (Genbank–NCBI) and USDA110 and USDA6 strains. They presented 100% of similarity in nucleotide sequences, showing a high level of evolutionary preservation within the species (unreported data). This lack of polymorphism between USDA110 and USDA6 strains identified for aapP gene, and the low level of polymorphism identified for aapP gene can be attributed to the nucleotide sequence preservation during the evolutionary process of Bradyrhizobium genus.
In future studies, the sequencing of these genes and the comparison of nucleotide sequences among B. japonicum strains analyzed in our studies will allow identifying possible polymorphisms of one or few nucleotides.
Conclusion
The difficulty to obtain amplicons by PCR of nopP , aapP, ilvD and bll3106 genes indicate that nucleotide sequences of genes of B. japonicum USDA110 strains utilized to build primers are distinct from the other studied strains. PCR-RFLP analyses of aapP gene with the utilization of three restriction enzymes for USDA110 strain presented a different band profile from the other studied strains with two or three utilized enzymes, indicating that USDA110 does not belong to B. japonicum species. Through PCR-RFLP analysis of aapP gene it was possible to confirm the presence of two copies of this gene in S204 strains, suggesting that there is a greater efficiency of nitrogen biological fixation for this strain.
Nucleotide sequences of aapP gene and nopP gene showed to be highly preserved through PCR-RFLP analysis, which explains the lack of polymorphism among the rhyzobacteria utilized in this study. Thus, the detection of polymorphisms for these genes in these species will only be possible with the utilization of comparative analyses (similarity) between these sequences which will point out differences in only one or few nucleotides.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgments
The authors thank Universidade Paranaense, Fundação Araucária, Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for their support.
About the Authors
Corresponding Author
L.D. Paccola-Meirelles
Programa de Pós-Graduação em Biotecnologia Aplicada à Agricultura, Universidade Paranaense, Umuarama, PR, Brasil
- Email:
- luziadoretto@prof.unipar.br
References
- Barcellos FG, Batista JSS, Menna P, Hungria M (2009). Genetic differences between Bradyrhizobium japonicum variant strains constrasting N2-fixation efficiency revealed by representational difference analysis. Arch. Microbiol. 191: 113-122. https://doi.org/10.1007/s00203-008-0432-0
- Bortolan S, Barcellos FG, Marcelino FC, Hungria M (2009). Expressão dos genes nodC, nodW e nopP em Bradyrhizobium japonicum estirpe CPAC 15 avaliada por RT-qPCR. Pesqui. Agropecu. Bras.44: 1491-1498. https://doi.org/10.1590/s0100-204x2009001100017
- Chueire LMO, Bangel EV, Mostasso FL, Pedrosa FO, et al. (2003). Classificação taxonômica das estirpes de rizóbio recomendadas para as culturas da soja e do feijoeiro baseada no seqüenciamento do gene 16s rRNA. Rev. Bras. Ciênc. Solo. 27: 833-840. https://doi.org/10.1590/s0100-06832003000500007
- Delamuta JR, Ribeiro RA, Ormeño-Orrillo E, Mello IS, et al. (2013). Polyphasic evidence supporting the reclassification of Bradyrhizobium japonicum group Ia strains as Bradyrhizobium diazoefficiens sp. nov. Int. J. Syst. Evol. Microbiol. 63: 3342-3351. https://doi.org/10.1099/ijs.0.049130-0
- Hungria M, Campo RJ, Mendes IC (2007). A importância do processo de fixação biológica do nitrogênio para a cultura de soja: componente essencial para a competitividade do produto brasileiro. Londrina: Embrapa Soja, 80p. (Embrapa Soja. Documentos, 283).
- Hungria M, Boddey LH, Santos MA, Vargas MAT (1998). Nitrogen fixation capacity and nodule occupancy by Bradyrhizobium japonicum and B. elkanii strains. Biol. Fertil. Soils. 27: 393-399. https://doi.org/10.1007/s003740050449
- Kalendar R, Lee D, Schulman AH (2009). Fast PCR software for PCR primer and probe design and repeat search. Genes, Genomes and Genomics. 3: 1-14.
- Kaneko T, Nakamura Y, Sato S, Minamisawa K, et al. (2002). Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Research. 9: 189-197. https://doi.org/10.1093/dnares/9.6.225
- Ribeiro RA, Barcellos FG, Thompson FL, Hungria M (2009). Multilocus sequence analysis of Brazilian rhizobium microsymbionts of common bean (Phaseolus vulgaris L.) reveals unexpected taxonomic diversity. Res. Microbiol. 160: 297-306. https://doi.org/10.1016/j.resmic.2009.03.009
- Santos MA, Nicolás MF, Hungria M (2006). Identificação de QTL associados à simbiose entre Bradyrhizobium japonicum, B. elkanii e soja. Pesqui. Agropecu. Bras. 41: 67-75. https://doi.org/10.1590/s0100-204x2006000100010
- Souza JAM (2006). Perfil transcricional de Bradyrhizobium elkanii SEMIA 587 in vitro e em simbiose com soja (Glycine max L.) através de microarranjo de DNA. 2006. 150 f. Doctoral thesis, Faculdade de Ciências Agrárias e Veterinárias, UNESP, Jaboticabal.
- Vincent JM (1970). A manual for the practical study of root-nodule bacteria. International Biological Program Hand- book 15. Blackwell Scientific Publications, Oxford, England. https://doi.org/10.2307/2402718
Keywords:
Download:
Full PDF- Share This