Cloning and functional characterization of the HbSYR1 gene encoding a syntaxin-related protein in Tibetan hulless barley (Hordeum vulgare L. var. nudum HK. f.)
Received: November 01, 2016
Accepted: August 31, 2017
Published: June 22, 2016
Genet.Mol.Res. 16(3): gmr16038909
DOI: 10.4238/gmr16038909
Abstract
Tibetan barley is a staple food for the natives of Qinghai-Tibet Plateau. Drought causes a reduction in barley production. In this study, the full-length cDNA of a gene encoding a syntaxin-associated protein was cloned from the leaves of a drought-resistant variety of barley, “Himalaya 10”; its expression was evaluated during drought stress and rehydration via real-time PCR. The cloned HbSYR1 cDNA sequence was 1300 bp in length, and included an 840-bp open reading frame that encoded 279 amino acids. Sequence analysis predicted the molecular weight of the encoded protein to be 42.08 kDa, with an isoelectric point of 4.98. ScanProsite analysis showed that the HbSYR1 protein contained a SNARE family characteristic motif, five casein kinase II phosphorylation sites, two N-glycosylation sites, four protein kinase C phosphorylation sites, and two N-myristoylation sites. The TMHMM prediction program indicated that the protein does not contain a transmembrane transfer ribbon. According to the SignalP 3.0 server, this protein does not contain a signal peptide, and is not a secretory protein. Instead, this protein was suggested to be localized in the cytoplasm, as predicted by the protein subcellular localization prediction tool (PSORT). Our results indicated that HbSYR was induced by drought stress and rehydration, and was determined to be a key gene for drought resistance and water retention in barley.
Introduction
Drought is one of the most devastating natural disasters in the world. It can lead to abiotic stress, which affects crop growth and yield. It also causes a series of physiological and metabolic changes in plants, including growth inhibition, irreversible damages, and even death. During its growth and development, the jointing stage of the Tibetan hulless barley coincides with the non-rainy season of Tibet, and is therefore often subjected to drought stress. As a result, it is of great importance to breed drought-resistant varieties of this wheat, and explore mechanisms of drought resistance to improve its yield and quality.
Drought resistance is a very complex trait controlled by multiple genes. It involves signal transduction and transcription of regulatory, protection, defense, and stress tolerance genes (Wang et al., 2003). Furthermore, drought resistance is closely associated with a variety of physiological and biochemical reactions, including osmotic adjustments, anti-oxidation, photosynthesis, alkaloid metabolism, nutrient transport and distribution, presence of plant hormones, as well as protein metabolism and dehydration protection (Fan et al., 2006). A major method of cell-to-cell interaction is via vesicle transport, which is vital for drought resistance in crops. A syntaxin-related protein is a protein subfamily belonging to the multifunctional SNARE protein family. It is vital to many physiological processes in plants, such as vesicle-mediated integration processes and signaling pathways (Feng et al., 2009), as well as responses to biotic and abiotic stress. Geelen et al. (2002) determined that the SNARE protein NtSyr1 in tobacco controlled the correct positioning of K+ channel proteins in the plasma membrane, and affects the plants’ responses to the abscisic acid (ABA) hormone via vesicular transport. Zhu et al. (2002) found that deletion of the AtSYP61 gene in Arabidopsis resulted in an increase in salt and osmotic stress sensitivity. Furthermore, expression of NPSN11 (a novel plant-specific SNARE) was increased under oxidative stress, and was found to be significantly reduced during salt and osmotic stress (Lipka et al., 2005; Bao et al., 2008). In addition, NtSyp121 (syntaxin-related protein NtSyr1) in tomatoes may regulate vesicle transport to the plasma membrane, mainly by regulating membrane fusion in ABA-dependent and independent stress response pathways (Geelen et al., 2002; Collins et al., 2003). These findings indicated that AtVAMP71/AtVAMP7C in v-SNAREs (vesicle SNAREs) are not only able to play protective roles during drought stress, but also contribute to ABA-mediated stomatal closures (Leshem et al., 2010). Yuan et al. (2015) cloned and characterized the HbSINA4 gene, and found it to be upregulated in Tibetan hulless barley during drought stress. However, there is still little information regarding the functions of the HbSYR1 gene.
Based on information from previous studies (Zeng et al., 2012), we cloned the fulllength barley syntaxin-related protein gene (HbSYR1) using reverse transcription polymerase chain reaction (RT-PCR). Preliminary bioinformatic analysis and investigation of HbSYR1 expression patterns under drought stress provided the basis for further exploration with respect to drought stress during plant seeding stages.
Materials and Methods
Materials
The drought-resistant barley, “Himalaya 10”, was provided by the Tibet Academy of Agriculture and Animal Husbandry Sciences.
Methods
Drought stress treatment in barley
A selection of healthy potted barley seedlings were divided into nine groups, with three seedlings per group. The nine treatments were as follows: 33.4, 27.5, 21.1, 15.5, 9.8, and 4.8% water holding caPacIties in the soil were designated as C1, C2, C3, C4, C5, and C6, respectively. Groups C7, C8, and C9 received 2, 4, and 8 h of rehydration, respectively, while the absolute water holding caPacIty in the supplemented soil was between 4.8 and 33.4%. Leaves were obtained from the seedlings, and were immediately placed in liquid nitrogen and stored at -80°C for further analysis. Group C4 was designated as the control.
HbSYR1 cloning
Based on transcriptome sequencing data from barley leaves at different stages of drought stress and rehydration, the specific primers for HbSYR1 (FP: 5'-CGGTACCATTTTGC AGGATTCATTTGAGCT-3'; RP: 5'-CACTAGTCCAGGTCGTGCTCAAGAGTTACT-3') were designed using the Primer Premier 5.0 software (Premier Biosoft Interpairs, Palo Alto, CA, USA). The first strand of cDNA was reverse transcribed from RNA, and was used as a template for the upstream fragment. The 20-μL reaction mixture contained 0.2 μL TaqDNA polymerase (TaKaRa, 5 U/μL), 2 μL 10X PCR reaction buffer (TaKaRa), 1.6 μL dNTP (10 mM), 0.8 μL upstream and downstream primers (10 μM for each), 2 μL cDNA, and 12.6 μL sterile water. Amplification was carried out in a S-1000 Thermal Cycler, and the reaction conditions were as follows: denaturation at 94°C for 4 min, followed by 34 cycles of denaturation at 94°C for 50 s, annealing at 51°C for 45 s, extension at 72°C for 1 min, and finally extension at 72°C for 10 min. The target products were used for cloning and sequencing.
Bioinformatic analysis
The HbSYR1 gene was analyzed with the following databases and softwares: NCBI (http://ncbi.nlm.nih.gov/), ExPAsy (http://www.expasy.org/tools/pi_tool.html), TargeP (http://www.cbs.dtu.dk/services/SignalP), PSORT (http://psort.ims.u-tokyo.ac.jp/form.html), and TMHMM (http://www.cbs.dtu.dk/services/TMHMM/), as well as other network resources.
The HbSYR1 amino acid sequence homology alignment and phylogenetic analysis were carried out using the MEGA5.1 software (Tamura et al., 2011).
Real-time PCR analysis
Real-time PCR analysis was performed in accordance with the SYBR Premix Ex TaqTM kit manual introductions (http://www.takara.com.cn). Quantitative PCR primers designed by cDNA sequence of HbSYR1 was as follows: FP: 5'-TTGAAGGAACGGATGGACGAT-3'; RP: 5'-TTGCTCACTCCTCCCTGTCTCA-3'. β-Tublin (FP: 5'-CCAAGTTCTGGGAGGTGATC TG-3'; RP: 5'-TTGTAGTAGACGTTGATGCGCTC-3') was used as the reference gene. Gene expression was extrapolated from the double standard curve, and was calculated as previously described by Zhang et al. (2005).
Gene cloning and functional verification
The barley stripe mosaic virus (BSMV) used, “Himalaya 10”, was composed of three RNA chains: α, β, and γ. The resulting BSMV recombinant viral vector contained α, β, γ, and γ-PDS. The above materials were provided by the Tibet Academy of Agriculture and Animal Husbandry Sciences, China.
Gene fragment amplification via PCR was carried out with 20 μL reaction mixture. The reaction protocol was as follows: denaturation at 94°C for 4 min; 34 cycles of denaturation at 94°C for 50 s; annealing at 51°C for 45 s; extension at 72°C for 1 min; final extension at 72°C for 10 min. PCR products were verified by electrophoresis on a 1% agarose gel, and the target gene fragments were recovered from the gel.
The BSMV γ vector and target gene fragment were digested with NotI and PacI, respectively. The 50-μL digestion mixture, containing 5 μL 10X buffer, 0.5 μL 10X BSA, 42.5 μL BSMV γ vector (target gene fragment), 2 μL NotI (PacI), was then incubated at 37°C for 12 h. The complete digestion products were purified and recovered using a TaKaRa purification kit. BSMV γ was ligated with exogenous DNA at 16°C for 15 h using a TaKaRa DNA ligase kit. The ligation mixture (20 μL) included 10 μL ligase buffer, 7 μL exogenous DNA, and 3 μL BSMV γ vector. The above ligation product was transformed into an Escherichia coli DH5α competent cell, and was incubated on ampicillin resistance plates. PCR was used to verify the positive clones for correct sequence and direction of the target gene. The plasmids were stored at -20°C.
The vectors α and γ, and the plasmid γ-target genes were linearized using MluI digestion. The β vector was linearized via SpeI digestion, which was carried out overnight at 37°C with 100 μL reaction mixture. Products were purified using a TaKaRa DNA purification kit, and 3 μL purified product was subjected to electrophoresis. The remainder of the products were stored at -20°C for future use.
The capped RNA was synthesized using an in vitro transcription system from Promega mMESSAGE Mmachine High Yield Capped RNA Transcription Kit. The in vitro transcription system contained 4 μL T7 5X buffer, 6 μL rNTP Mix (GTP 2 mM, else 25 mM), 6.5 μL DNA, 1.5 μL 40 mM Ribo m7G, 2 μL Enzyme Mix (T7), and 6.5 μL ddH2O. The reaction was carried out at 37°C for 4 h, and the products were then stored at -70°C.
During friction inoculation, the BSMV recombinant virus was inoculated on the third leaf during the trefoil stage of the barley; the PDS gene was used as a marker gene, and moisture was applied for 24 h. The inoculated barley plants were then placed on a 16/8 h light/ dark cycle, and cultured in a greenhouse at 25°C. Seedlings with non-inoculated virus were used as the controls. The gene silencing efficiency was analyzed on the 14th day following inoculation. Phenotypes of the seedlings that underwent drought stress with a 4.8% soil water holding capability were examined. When plants recovered up to 33.4% of soil water-holding capability, leaf samples were taken at 2 and 4 h after rehydration, respectively. The samples were placed in liquid nitrogen, and stored at -80°C for further analysis.
Results
HbSYR1 gene cloning and bioinformatic analysis
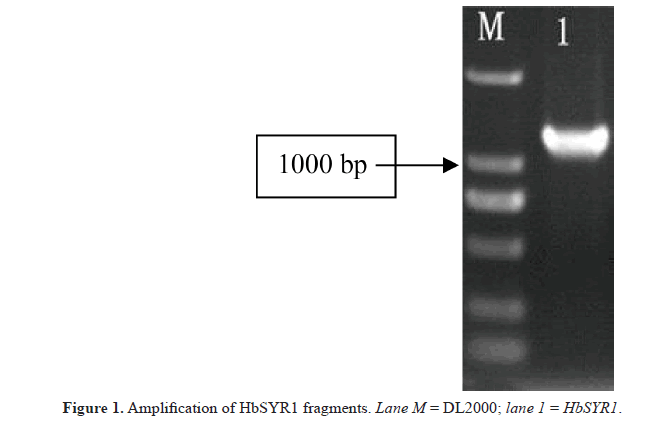
In this study, specific primers were designed for a cDNA sequence encoding the SYR gene in barley. The 1300-bp cDNA sequence was amplified from the leaves of barley seedlings (Figure 1). The length of the 5’-UTR and 3’-UTR were 260 and 200 bp, respectively. The length of the gene open reading frame (ORF) was 840 bp, which encoded a 279-amino acid polypeptide, as demonstrated by the ORF finder program on the NCBI website (Figure 2). The molecular weight of the encoded protein was predicted to be 42.08 kDa, and the isoelectric point was 4.98. ScanProsite analysis showed that the HbSYR1 protein contained a SNARE family characteristic motif (182 to 244 amino acids sites), five casein kinase II phosphorylation sites (38 to 41,75 to 78, 97 to 100, 152 to 155, and 181 to 184 amino acids sites), two N-glycosylation sites (84 to 87 and 229 to 232 amino acids sites), four protein kinase C phosphorylation sites (86 to 88, 127 to 129, 159 to 161, and 251 to 253 amino acids sites), and two N-myristoylation sites (92 to 97 and 239 to 244 amino acids sites). The TMHMM predicted that the protein did not contain a transmembrane transfer ribbon. The SignalP 3.0 server indicated that no signal peptide sequence was present on the protein, and it was not a secretory protein. The PSORT subcellular localization software predicted that HbSYR1 protein was localized in the cytoplasm.
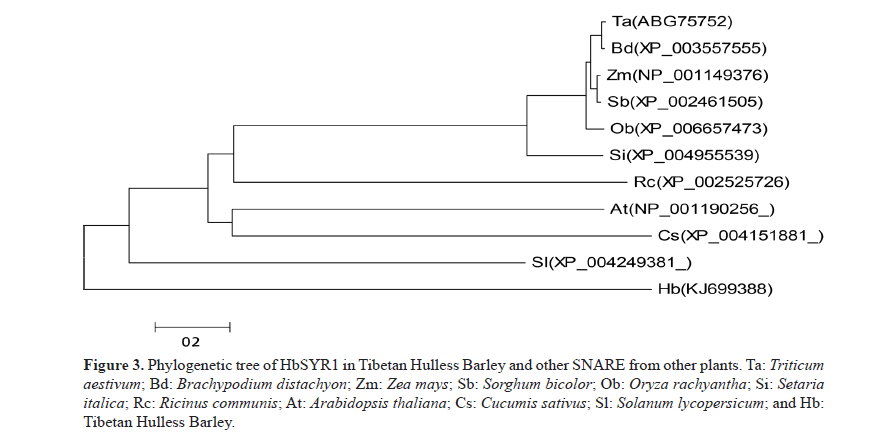
BLASTn analysis indicated that the full-length cDNA was similar to SYR gene sequences of Triticum aestivum, Brachypodium distachyon, Setaria italica, Zea mays, Sorghum bicolor, Oryza brachyantha, Aegilops umbellulata, Lycopersicon esculentum, and Arabidopsis thaliana, which were at 99, 97, 92, 92, 92, 90, 84, 70, and 64% homology, respectively. Analysis of the deduced amino acid sequences alignment (Figure 3) showed that its sequence were similar to the SYR sequences of T. aestivum, B. distachyon, S. italica, Z. mays, S. bicolor, O. brachyantha, and A. thaliana, which were at 99.0, 97.0, 93.0, 93.0, 90.0, 90, 70, and 70% homology, respectively. A phylogenetic tree constructed based on amino acid sequence showed that evolution of the amino acids was consistent with that of other plants. In addition, a close relationship was found between the amino acid sequences of this protein and other SNARE proteins in T. aestivum, S. italica, and B. distachyon. However, there was a low sequence similarity to those of O. brachyantha, S. bicolor, and A. thaliana. Therefore, this SNARE gene inferred from its full-length cDNA sequence in barley was named HbSYR1 (GenBank accession No.: KJ699388).
Figure 3: Phylogenetic tree of HbSYR1 in Tibetan Hulless Barley and other SNARE from other plants. Ta: Triticum aestivum; Bd: Brachypodium distachyon; Zm: Zea mays; Sb: Sorghum bicolor; Ob: Oryza rachyantha; Si: Setaria italica; Rc: Ricinus communis; At: Arabidopsis thaliana; Cs: Cucumis sativus; Sl: Solanum lycopersicum; and Hb: Tibetan Hulless Barley.
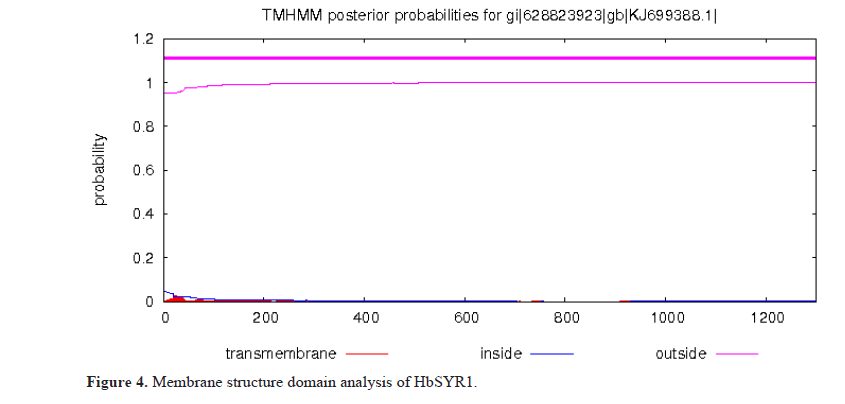
According to the analysis and prediction of the transit peptide by TargetP, it was determined that HbSYR1 contained cytoplasmic proteins without transit peptides. In addition, as predicted by the online software TMHMM, it did not contain any transmembrane regions (Figures 4 and 5).
Expression pattern analysis of the HbSYR1 gene
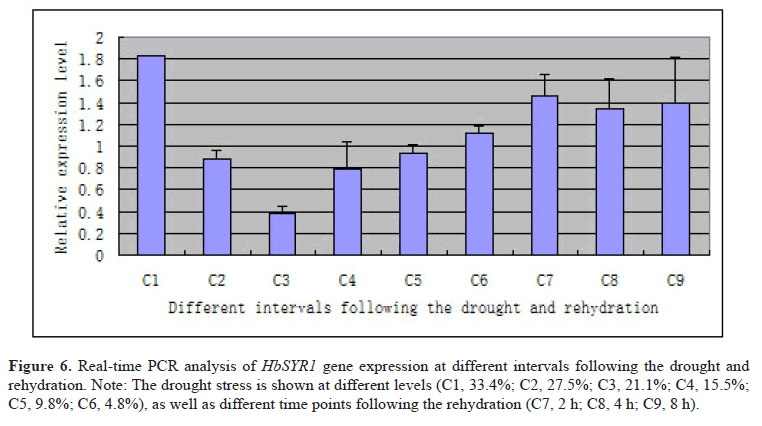
In order to explore the gene expression pattern of HbSYR1, real-time PCR was performed at nine different time points in barley leaves undergoing drought stress and rehydration. The results (Figure 6) showed that the HbSYR1 gene was significantly upregulated when absolute water content in the soil was at 33.4%, and sharply declined with decreasing water content. It was again upregulated when the absolute soil water content was recovered to that required for crop growth. Following drought stress, the HbSYR1 gene was significantly upregulated when absolute water content was reduced to 4.8%, and showed highest expression at 2 h after rehydration. As rehydration time was extended, its expression level declined and trended towards stabilization. This suggested that the HbSYR1 gene may be involved in regulation of the waterlogging and drought stress-associated signal transductions.
Figure 6: Real-time PCR analysis of HbSYR1 gene expression at different intervals following the drought and rehydration. Note: The drought stress is shown at different levels (C1, 33.4%; C2, 27.5%; C3, 21.1%; C4, 15.5%; C5, 9.8%; C6, 4.8%), as well as different time points following the rehydration (C7, 2 h; C8, 4 h; C9, 8 h).
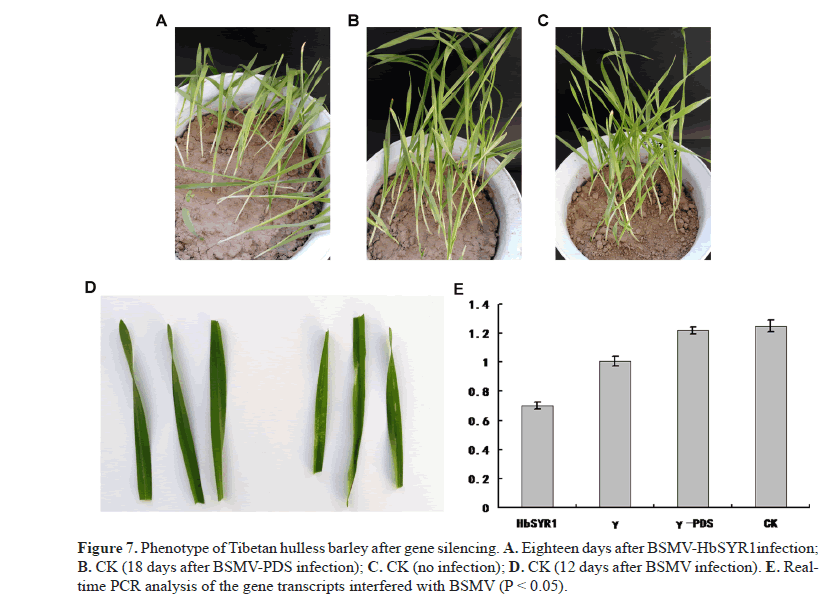
Gene silencing of HbSYR1 and real-time quantitative PCR analysis
Virus-induced gene silencing was used to degrade post-transcriptional mRNAs, which resulted in transcriptional silencing of the target gene. Total RNA in the barley leaves was extracted and real-time quantitative PCR was performed to confirm gene silencing. Following gene silencing, drought stress treatment was carried out on the 14th day post-inoculation. Seedlings with BSMV-HbSYR1 gene silencing showed distinct phenotypic variation when water-holding caPacIty in the soil was reduced to 4.8%, as compared to the control (Figure 7A). Drought tolerance was difficult to achieve due to severe wilting symptoms such as chlorosis and yellowing (Figure 7B) 12 days after transfection with BSMV. The degree of wilting in plant inoculated with BSMV-PDS was similar to that of controls when the water holding caPacIty in the soil was reduced to 4.8% (Figure 7C), with the controls showing slightly less wilting (Figure 7D). Barley plants with silenced HbSYR1 gene showed less drought tolerance overall, suggesting that this gene plays a role in the drought resistance of the barley.
Real-time quantitative PCR results showed that partial silencing of the HbSYR1 gene was achieved, with a 44.8% reduction in expression level as compared with the control (Figure 7E).
Discussion
SNARE proteins in plants not only play a role in plant growth and development, but are also important during biotic and abiotic stress (Feng et al., 2009). For example, the AtVAMP71 family of SNARE proteins in Arabidopsis were found to control H2O2 vesicle transport and regulate reactive oxygen species accumulation and distribution in the cells. Therefore, they play a role in salt stress tolerance plants. At the same time, the AtVAMP71 gene is also important for adjusting the opening and closing of the plant stomata, and controlling ABA content during drought stress (Leshem et al., 2010). In addition, VAM3 gene mutation in Arabidopsis roots increased Na+/H+ transporter gene expression, leading to increased Na+ content in the roots. However, it reduced Na+ content in the leaves, and ultimately improved salt tolerance in plants (Sato et al., 1997). Sun et al. (2013) suggested that GsSNARE1 was expressed in different tissues, and was induced by salt and drought stress to regulate stress tolerance in recombinant bacteria. Zhu et al. (2002) found that OSM1/SYP61 genes in Arabidopsis were associated with osmotic stress and stomatal closure, and suggested that SNARE proteins also participate in signal transduction of the abiotic stress dependent and independent induction of ABA. SNARE proteins in plants also interact with the ion channel proteins. Geelen et al. (2002) reported that the homologous genes NtSyr1 of synaptic fusion protein SYP121 in tobacco regulate K+ channel protein in intracellular transport, and its expression is induced by ABA, salt-stress, drought stress, and injuries. Deletion of NtSyr1 affected the correct positioning of K+ channel proteins in the plasma membrane, and inhibited ion channel responses to ABA. Sokolovski et al. (2008) also found that SNARE proteins in plants may indirectly regulate ABA signals to the K+ and Cl- channels via the Ca2+ signaling pathway. In this study, we found that HbSYR1 is important for drought resistance and water retention in Tibetan barley. This is consistent with studies conducted by Geelen et al. (2002) , Leshem et al. (2010), and Sun et al. (2013), which suggested that HbSYR1 plays key roles in biotic and abiotic stress resistance.
It is known from previous studies that SNARE proteins are mainly involved in vesicle transport. Furthermore, these proteins can promote formation of cell plates in plants, interact with ion channel proteins, improve plant disease resistance, and participate in the gravitropism effects in plants (Bao et al., 2005). However, it is not yet clear how SNARE proteins interact with other factors to regulate vesicle transport, how they allow plants to adapt to extreme conditions, or how they coordinate signal transduction pathways through the entire plant. In this study, the SNARE regulation related genes, HbSYR1, were cloned. These genes are associated with the signal transductions during waterlogging and drought stress in barley, and are important drought stress genes. However, the specific mechanism underlying the function of the HbSYR1 gene is not yet clear. Further exploration is required to determine whether it is similar to the NtSyr1 gene in tobacco, and whether signal transduction during abiotic stress is dependent on the ABA through its interactions with ion channel proteins.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
Research supported by the following funding sources: the Tibet Autonomous Region Financial Special Fund (#2015CZZX001), the Tibet Autonomous Region Financial Special Fund (#2017CZZX001/02), and the Tibet Autonomous Region Financial Special Fund (#2014CZZX001).
About the Authors
Corresponding Author
N. Tashi
Tibet Academy of Agriculture and Animal Husbandry , Lhasa, China
- Email:
- nima_zhaxi@sina.com
References
- Bao YM, Wang ZF and Zhang HS (2005). Structure and function of SNAREs in plant. Chin. Bull. Bot. 22: 715-722.
- Bao YM, Wang JF, Huang J and Zhang HS (2008). Cloning and characterization of three genes encoding Qb-SNARE proteins in rice. Mol. Genet. Genomics 279: 291-301. http://dx.doi.org/10.1007/s00438-007-0313-2
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, et al. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973-977. http://dx.doi.org/10.1038/nature02076
- Fan M, Jin L and Li C. (2006). Research progress on the Mechanism of Drought Resistance in Potato. Chin. Potato 20: 101-107.
- Feng H, Chen C, Wang YQ, Qiu JL, et al. (2009). Plant SNAREs and their biological functions. Yi Chuan 31: 471-478. http://dx.doi.org/10.3724/SP.J.1005.2009.00471
- Geelen D, Leyman B, Batoko H, Di Sansebastiano GP, et al. (2002). The abscisic acid-related SNARE homolog NtSyr1 contributes to secretion and growth: evidence from competition with its cytosolic domain. Plant Cell 14: 387-406. http://dx.doi.org/10.1105/tpc.010328
- Leshem Y, Golani Y, Kaye Y and Levine A (2010). Reduced expression of the v-SNAREs AtVAMP71/AtVAMP7C gene family in Arabidopsis reduces drought tolerance by suppression of abscisic acid-dependent stomatal closure. J. Exp. Bot. 61: 2615-2622. http://dx.doi.org/10.1093/jxb/erq099
- Lipka V, Dittgen J, Bednarek P, Bhat R, et al. (2005). Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310: 1180-1183. http://dx.doi.org/10.1126/science.1119409
- Sato MH, Nakamura N, Ohsumi Y, Kouchi H, et al. (1997). The AtVAM3 encodes a syntaxin-related molecule implicated in the vacuolar assembly in Arabidopsis thaliana. J. Biol. Chem. 272: 24530-24535. http://dx.doi.org/10.1074/ jbc.272.39.24530
- Sokolovski S, Hills A, Gay RA and Blatt MR (2008). Functional interaction of the SNARE protein NtSyp121 in Ca2+ channel gating, Ca2+ transients and ABA signalling of stomatal guard cells. Mol. Plant 1: 347-358. http://dx.doi. org/10.1093/mp/ssm029
- Sun MZ, Sun XL, Yu QY, Wei JI, et al. (2013). Functional analysis of a stress-induced SNARE gene GsSNARE1 in response to salt and drought stresses. China Agric. Sci. 46: 2191-2200.
- Tamura K, Peterson D, Peterson N, Stecher G, et al. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731-2739. http://dx.doi.org/10.1093/molbev/msr121
- Wang W, Vinocur B and Altman A (2003). Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218: 1-14. http://dx.doi.org/10.1007/s00425-003-1105-5
- Xu YY, Wang MG and Jia JF (2001). Selection and identification of salt tolerant line of sainfoin from the seeds of first post-flight plants. Shi Yan Sheng Wu Xue Bao 34: 11-15.
- Yuan HJ, Luo XM, Nyima TS, Wang YL, et al. (2015). Cloning and characterization of up-regulated HbSINA4 gene induced by drought stress in Tibetan hulless barley. Genet. Mol. Res. 14: 15312-15319. http://dx.doi.org/10.4238/2015. November.30.7
- Zeng X, Tashi N and Yang J (2012). Construction of SSH library under drought stress, and the analysis of drought-induced gene expression in Tibetan barley varieties Himalayan No. 10. J. Wheat Crops 32: 640-647.
- Zhang C, Chee H and Huang X (2005). Establishment of a novel simple fluorescence relative quantitative real-time RTPCR method. Prog. Biochem. Biol. Phys. 32: 883-888.
- Zhu J, Gong Z, Zhang C, Song CP, et al. (2002). OSM1/SYP61: a syntaxin protein in Arabidopsis controls abscisic acid-mediated and non-abscisic acid-mediated responses to abiotic stress. Plant Cell 14: 3009-3028. http://dx.doi. org/10.1105/tpc.006981
Keywords:
Download:
Full PDF- Share This