Detection of sequences related to biosynthesis of antifungal metabolites and identification of a PhzF-like gene in a Pseudomonas rhizobacterium of Piper tuberculatum
Received: May 25, 2018
Accepted: June 14, 2018
Published: July 26, 2018
Genet.Mol.Res. 17(3): http://dx.doi.org/gmr18035
DOI: http://dx.doi.org/10.4238/gmr18035
Abstract
Our previous studies identified a Pseudomonas putida (Pt12 isolate) associated with roots of Piper tuberculatum Jacq., a Piperaceae occurring in the Amazon region with known resistance to infection by Fusarium solani f. sp. piperis, the causal agent of root rot disease in black pepper (Piper nigrum L.). This Pt12 isolate was able to inhibit in vitro growth of this fungus 39%. However, the inhibition mechanisms were unknown. Here, we searched for genes coding enzymes involved in the biosynthesis of compounds with potential antifungal activity in the Pt12 isolate, such as phenazine, pyoluteorin, 2,4-diacetylphloroglucinol and hydrogen cyanide. We found sequences potentially related to biosynthesis of phenazine (PhzF) based on DNA sequencing and comparative sequence analyses using the GenBank DataBase. PCR and nested PCR assays were used to isolate a 789-bp sequence coding for a deduced protein with 262 amino acid residues with high identity with a PhzF family phenazine biosynthesis protein from P. putida (WP_046785327.1), P. plecoglossicida (WP_041505738.1) and P. parafulva (WP_039578870.1). Molecular modeling and molecular dynamic simulations revealed that putative PhzF of Pt12 isolate could stably interact with substrate DHHA (Trans-2, 3-dihydro-3- hydroxyanthranilic acid) through some amino acid residues that are similar to conserved amino acids critical to the catalytic activity of a well-characterized PhzF protein produced by Pseudomonas fluorescens.
Introduction
Endophytic bacteria are considered microorganisms that are able to colonize the internal tissues of plants, benefiting the host through the production of plant growth promoting substances, such as phytohormones, and/or by fixing nitrogen from the atmosphere and/or solubilization of phosphates (Glick, 2012). In addition, several endophytic bacteria have been found aiding plants against pathogens through plant disease suppression (Glick, 2012). Endophytic bacteria able to colonize plant roots and to promote plant growth are known as plant growth-promoting rhizobacteria (PGPR) (Beneduzi et al., 2012). Among PGPR, Pseudomonas bacteria have been useful for the control of plant diseases caused by many pathogens, including fungi, such as Macrophomina phaseolina (Vanitha and Ramjegathesh, 2014), Fusarium spp. (Sarhan and Shehata, 2014; Nascimento et al., 2015), and Glomerella tucumensis (Hassan et al., 2011).
Pseudomonas mechanisms to suppress plant diseases can include induced systemicresistance, where plants in contact with endophytic bacteria show enhanced expression of genes involved in a defense response when infected by pathogens (Verhagen et al., 2010). Also, Pseudomonas bacteria can produce substances affecting infection by pathogens, including siderophores (Glick, 2012) and metabolites, such as phenazine, pyoluteorin, 2,4-diacetylphloroglucinol (DAPG), pyrrolnitrin and hydrogen cyanide (HCN) (Ramette et al., 2003; Keshavarz-Tohid et al., 2017).
The potential of bacterial metabolites to control phytopathogens has been evidenced by many studies. For instance, Puopolo et al. (2013) reported the potential of phenazines against 11 phytopathogenic fungi. In alfalfa, a P. putida producing HCN was able to attenuate infection by F. solani (Sarhan and Shehata, 2014). Likewise, a pyoluteorin produced by P. putida decreased symptoms of red root rot disease caused by Glomerella tucumensis in sugar cane (Hassan et al., 2011). In this context, Pseudomonas bacteria withpotential to control phytopathogens have been characterized through detection of metabolites produced by them and/or by detection of bacterial genes related to biosynthesis of such compounds (Cornea et al., 2007; Liu et al., 2006; Hassan et al., 2011; Zhang et al., 2018).
Among Pseudomonas spp. with potential to control phytopathogens, Nascimento et al. (2015) reported a Pseudomonas putida (Pt12 isolate) associated with roots of Piper tuberculatum Jacq., a Piperaceae occurring in the Amazon region with known resistance to infection by Fusarium solani f. sp. piperis (Albuquerque et al., 2001). This pathogen is the causal agent of root rot or fusariosis disease in black pepper (Piper nigrum L.) (Albuquerque et al., 1976), known as ‘the king of spices’, with great economic and social importance for Pará State, in northern Brazil, where this disease drastically affects its production. The Pará state participation in black pepper production is significant, corresponding to 63% of the 2016 Brazilian harvest, positioning this state as the largest regional and national producer. However, Brazilian black pepper production has been declining; production was reduced from 80,316 t in 2006 to 42,339 t in 2014 (IBGE, 2016). Much of this reduction is due to this disease. The Pt12 isolate showed potential to control root rot disease of black pepper, since it was able to inhibit the in vitro growth of F. solani f. sp. piperis 39% (Nascimento et al., 2015). However, the inhibition mechanisms are still unknown.
In attempt to find out how this isolate is able to inhibit fungal growth, we searched for gene sequences related to the biosynthesis of phenazine, pyoluteorin, pyrrolnitrin, DAPG and HCN in the Pt12 isolate. We found sequences potentially related to phenazine biosynthesis, according to DNA sequencing and comparative sequence analyzes in the GenBank Database. To date, phenazine and its derivatives, such as phenazine-1-carboxamide (PCN) and phenazine-1-carboxylic acid (PCA) have been considered the most potent antibiotic metabolites, exhibiting broad-spectrum activity against bacteria and fungi (Bilal et al., 2017). In Pseudomonas bacteria, at least seven genes constitute the operon for the phenazine biosynthetic pathway: PhzABCDEFG (Mavrodi et al., 1998, Mavrodi et al., 2001a; Bilal et al., 2017). The PhzF gene encodes the PhzF protein, a key enzyme in the conversion of DHHA (Trans-2, 3-dihydro-3-hydroxyanthranilic acid) in PCA, and contains one active site with the conserved catalytically critical residues E45, H74 and D208 (Parsons et al., 2004; Blankenfeldt et al., 2004, Mavrodi et al., 2010; Liu et al., 2015).
Here, we report the isolation of a 789-bp PhzF sequence of Pt12 isolate, coding for a deduced protein with 262 amino acid residues with high identity with PhzF family phenazine biosynthesis protein from P. putida (WP_046785327.1), P. plecoglossicida (WP_041505738.1) and P. parafulva (WP_039578870.1). Molecular modeling and molecular dynamic simulations showed that this putative PhzF of the Pt12 isolate stably interacted with the substrate DHHA through some amino acid residues, which are similar to conserved amino acids found in the active site of a well-characterized PhzF protein of Pseudomonas fluorescens.
Material and Methods
Genomic DNA of Pt12
Bacterial cells of P. putida Pt12 isolate obtained by Nascimento et al. (2015) were cultured in tryptone soybean medium (Himedia, India) and used for isolation of genomic DNA according to methodology described by the same authors. Genomic DNA samples were quantified using a Qubit fluorometer (Invitrogen, USA) and evaluated by electrophoresis in agarose gel stained with ethidium bromide. Samples containing 0.1-0.2 µg of genomic DNA were used in PCR assays.
Detection of sequences related to antifungal metabolites in Pt12
Sequences of genes related to biosynthesis of phenazine, pyoluteorin, pyrrolnitrin, DAPG and HCN were amplified by PCR assays using primers and conditions described in the literature and primers designed for this study. For DAPG (PhlD), we used B2BF/BPR4 primers described by McSpadden et al. (2001). Sequences for Pyoluteorin (PltB) were amplified using PltBf/PltBr and plt1/plt2 primers combinations reported by Mavrodi et al. (2001b). For Pyrrolnitrin (prnC), we used Pyr1 and Pyr2 primers reported by Allaire (2005), while for HCN (hcnBC) we used ACa and ACb primers according to Ramette et al. (2003). For Phenazine (PhzCD), PCA2a and PCA3b primers were used according to Raaijmakers et al. (1997). For Phenazine PhzF, PHEPUT-F1 (5’GCAGCGCATCGCCGAAGAGAACA3’) and PHEPUT-R1 (5’GTAACGGCGATGCCGCGCACAT3’) primers based on Accession JX843728.1 from GenBank were designed using the Vector NTI Advance version 11.1 (Invitrogen, USA). Primers for the 16S rDNA gene, used as a positive control, were the same used by Nascimento et al. (2015). For PhzF and 16S rDNA primers, we used 55oC for annealing temperature with the same PCR conditions reported by Nascimento et al. (2015).
After amplification, PCR products were analyzed by electrophoresis in agarose gels stained with ethidium bromide. DNA fragments with expected size were purified from the agarose gel using the Gel DNA Recovery kit (ZymoResearch, USA) and cloned into the pGEM Teasy vector (Promega, USA), following the manufacturer’s recommendations. Electrocompetent Escherichia coli, Top 10 strain (Invitrogen, USA) was transformed using an Electroporator 2510 (Eppendorf, Germany), following the manufacturer’s recommendations. Transformed bacteria were plated on Petri dishes containing LB Broth with agar (Sigma Aldrich, USA) supplemented with ampicillin (100 µg/mL). For screening of recombinant clones, we used X-GAL (5-bromo-4-chloro-3- indolyl-β-D-galactopyranoside) and IPTG (Isopropyl β-D-1-thiogalactopyranoside).
Amplification of the PhzF sequence of the Pt12 isolate
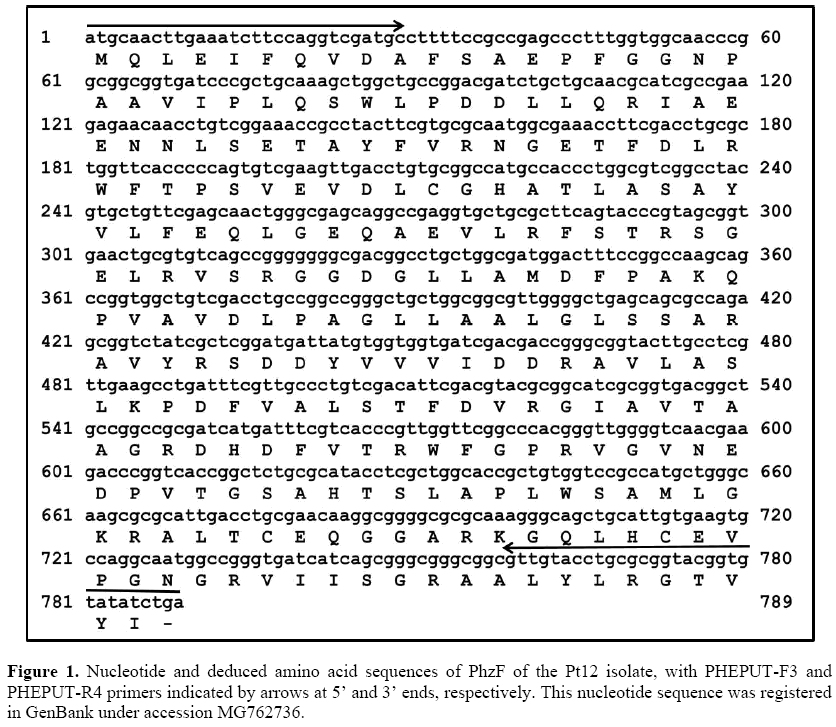
Sequences corresponding to PhzF ORF (Open Reading Frame) were amplified by PCR and nested PCR using primers specific for partial PhzF sequence of Pt12 isolate and primers based on PhzF sequence of P. putida strain DLL-E4, considered to be the closest lineage. PHEPUT-F3 (5’ATGCAACTTG AAATCTTCCAGGTCGATGc3’) and PHEPt12-R1 (5’CGATAGACCGCTCT GGCGCTGCTC3’) primers were used for amplification of 5’ end. For amplification of 3’ end we used nested PCR assays, with PHEPt12-F2 (5’TGTCGAAGTTGACCTGGTCGGCCAT3’) and PHEPUT-R4 (5’TCAGAT ATACACCGTACCGCGCAGGTACAAC3’) primers in the first PCR, and PHEPt12-F1 (5’AGCAGGCCGAGGTGCTGCGCT3’) and PHEPUT-R4 primers in the second PCR. PCR products were evaluated by electrophoresis and cloning into the pGEM Teasy vector, as described above.
DNA sequencing and sequence analyzes using computational tools
DNA sequencing was carried out using the Big Dye Terminator Kit and ABI 3130 Automatic Sequencer (Applied Biosystems, USA). First, the nucleotide sequences were qualitatively evaluated with Bioedit Software (Hall, 1999), with subsequent comparison to sequences from GenBank Database using BLAST tools from NCBI (National Center of Biotechnology Information). Bioedit Software (Hall, 1999) was also used to assemble the 5’ and 3’ sequences of PhzF gene, which were subsequently analyzed by similarity with the non-redundant GenBank protein database using the BLASTX algorithm (Altschul et al., 1997). Molecular weight and isoelectric point of the PhzF deduced amino acid sequence were predicted by ExPASy Proteomics Server (http://ca.expasy.org/tools/pi_tool.html). Bioedit Software (Hall, 1999) was also used in the multiple alignments of the PhzF protein sequences. The LALIGN tool from ExPASy was used to calculate the identity between the deduced amino acid sequence of PhzF of Pt12 isolate and PhzF proteins from GenBank. The nucleotide sequence for PhzF of the Pt12 isolate was registered in GenBank Database from NCBI.
Molecular modeling of the putative PhzF of Pt12 isolate
The PDB database was searched using the BLAST algorithm included in the server. The target sequence of PhzF of Pt12 was provided in FASTA format and the results were evaluated based on alignment score, e-value and resolution of the structure. The alignment of the template and the target sequences was performed by PROMALS 3D (Pei et al., 2008) considering their structural characteristics. MODELLER 9.16 (Webb and Sali, 2016) was used to build the structural models. A total of 100 models were generated, considering different conformations, and ranked by DOPE energy. The models were generated satisfying the spatial restrictions, such as bond lengths, bond angles, dihedral angles and interactions between non-bonded segments and then subjected to validation. The model’s stereochemical quality was evaluated using a Ramachandran plot generated in the MolProbity server (Davis et al., 2007). The quality of folding was determined by Verify 3D (Luthy et al., 1992) and QMEAN (Benkert et al., 2008). Finally, the Root Mean Square Deviation (RMSD) between the main chain of the template and the target was computed.
Molecular dynamics of putative PhzF protein and DHHA interaction
The H ++ server (http://biophysics.cs.vt.edu/index.php) was used to determine the protonation state of the protein considering a 7.0 pH level. System assembly, as well as the energy minimization steps, heating and molecular dynamics were performed using tools included in the AMBER 16 package. The gaff and ff14SB force fields were applied for ions and protein, respectively (Maier et al., 2015). Thus, TIP3P water molecules were placed in a 12Å octahedral box in each direction of the protein. In order to neutralize the system, Na+ ions were added. Thus, the topology files and parameters for the system were created. The assembled system was submitted to the energy minimization step by sander. The energy minimization was performed in five steps, four of these using 3000 cycles of steepest descent and 5000 cycles of conjugate gradients for each one; the heavy atoms were restrained by a harmonic potential of 1,000 Kcal/mol*Å2. In the last step, we used 5,000 cycles of steepest descent and 30,000 cycles of conjugate gradients and no restraints. The heating and equilibration stage was divided into 14 steps. The temperature was gradually increased, until it reached 300 K. Langevin Dynamics (thermostat) was employed with a collision frequency of 3.0 ps−1. A harmonic potential of 25 Kcal/mol*Å2 was employed in the initial steps and was turned off in step 13. The heating procedure lasted 650 picoseconds until step 13 and was performed using an NVT ensemble. Afterwards, a 2 nanosecond equilibration phase was employed in an NPT ensemble. The SHAKE algorithm was employed to restrict ligations vibration of all hydrogen atoms. The Particle Mesh Ewald method was used for calculating electrostatic interactions using a cutoff value of 10.0 Å. Thus, 100 ns MD was produced in an NVT ensemble, for each system. The cpptraj module was used to compute the Root Mean Square Deviation (RMSD) of trajectory, considering the heavy atoms of the main chain. MMPBSA.py Amber script (Genheden and Ryde, 2015) was used to estimate the affinity of HHA ligand to the protein using 5000 frames in a single trajectory protocol.
Results
Detection of sequences related to biosynthesis of antifungal metabolites in the Pt12 isolate
Our results showed the amplification of DNA fragments with expected size for pyoluteorin and phenazine based on electrophoresis in agarose gel stained with ethidium bromide. No expected DNA bands were detected by other combinations of primers, except for the 16S rDNA sequence used as a control. Besides electrophoresis in agarose gel, we also submitted the amplified fragments to DNA sequencing and comparative sequence analyses using the Blast-X tool from NCBI in order to confirm the amplification of target sequences. After these analyses, we confirmed DNA fragments potentially related to biosynthesis of phenazine only.
Regarding pyoluteorin, we used PltBf and PltBr primers described by Mavrodi et al. (2001b) based on pyoluteorin biosynthetic gene cluster from P. fluorescens Pf-5 (Accession AF081920 in GenBank) with the gene PltB for polyketide synthase, which contains the acyltransferase and acylcarrier protein domains, among others. Although a DNA band with an expected size showed 65% identity with the acyltransferase of Pseudomonas (WP_085531747), this sequence had no significant identity with a 792-bp sequence flanked by PltBf and PltBr primers within the polyketide synthase sequence (AF081920).
For phenazine, a 432-bp DNA fragment amplified using PHEPUT-F1/PHEPUT-R1 primers showed over 85% identity with PhzF proteins from Pseudomonas putida GB-1 (ABY99773.1) and Pseudomonas chlororaphis (SDS67984.1). In addition, sequence analyzes showed the 432-bp fragment as a partial sequence, where 429 bp corresponded to 143 aa, lacking 35 aa and 84 aa at the amino and carboxy-terminals, respectively, since the PhzF proteins homologues to the Pt12 PhzF are constituted of 262 aa.
Isolation and characterization of nucleotide and deduced amino acid sequences of PhzF of the Pt12 isolate
We performed PCR and nested PCR assays in order to amplify the sequence corresponding to the complete ORF for PhzF of the Pt12 isolate. For the 5’ end, we amplified a 432-bp fragment, while for the 3’ end a 595-bp fragment was amplified. After assembling 5’ and 3’ sequences, we obtained a complete ORF with 789 bp coding a deduced amino acid sequence with 262 aa (Figure 1).
Bioinformatics analyzes revealed that the putative PhzF protein of Pt12 isolate has a predicted molecular weight of 28 kDa and an isoelectric point of 5.0. Also, this protein showed significant identity with PhzF proteins available in GenBank (Table 1). Among them, the PhzF of Pt12 was 88.9% identical to the PhzF family phenazine biosynthesis protein of Pseudomonas putida (WP_046785327.1), and about 87% identical to the PhzF family phenazine biosynthesis protein from Pseudomonas plecoglossicida (WP_041505738.1) and Pseudomonas parafulva (WP_039578870.1). On the other hand, our sequence showed low identity with PhzF proteins from Pseudomonas fluorescens (AAC18905.1) and Pseudomonas aeruginosa (AAC64492.1).
| Accession | Organism | Identity (%) |
|---|---|---|
| W P_021782991.1 | M ultispecies: [Pseudom onas] | 89.3 |
| W P_046785327.1 | Pseudomonas putid a | 88.9 |
| W P_041505738.1 | P seudomonas plecoglossicida | 87.4 |
| AEJ14259.1 | Pseudomonas putida S16 | 87.4 |
| AGA74580.1 | P seudomonas putida H B3267 | 87.4 |
| AFO48928.1 | Pseudomonas putida DOT-T1E | 87.4 |
| W P_039578870.1 | Pseudomonas parafulva | 86.6 |
| AAC18905.1 | Pseudomonas fluorescen s 2-79 | 32 |
| AAC64492.1 | Pseudomonas. aeruginos a PAO1 | 31.4 |
Table 1: Identity between the deduced amino acid sequences of PhzF of the Pt12 isolate and PhzF proteins from GenBank using the LALIGN tool from ExPASy.
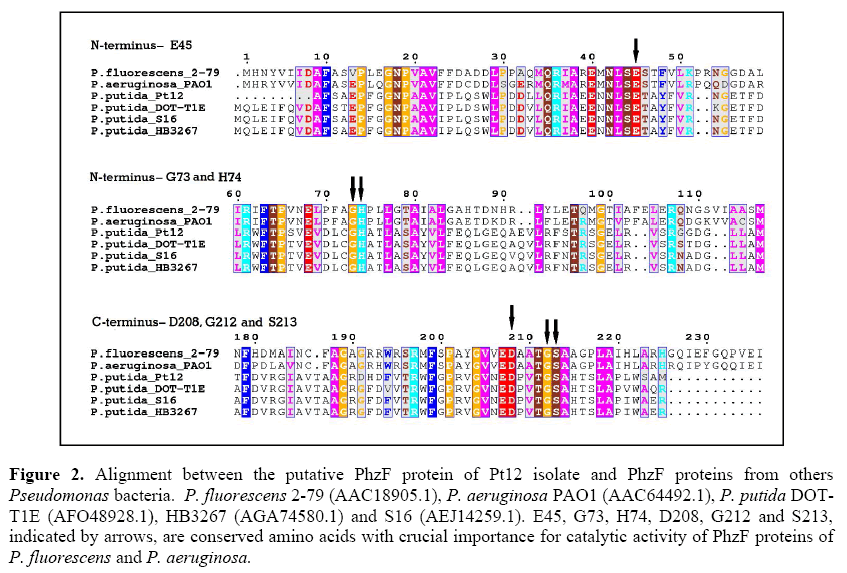
Figure 2 shows the alignment between the PhzF of Pt12 and PhzF proteins from other Pseudomonas bacteria, including P. fluorescens and P. aeruginosa with the conserved amino acids E45, H74 and D208, which are critical to their catalytic activity (Parsons et al., 2004; Mavrodi et al., 2010; Liu et al., 2015). Besides these three amino acids, the conserved G73, G212 and S213 (Parsons et al., 2004; Blankenfeldt et al., 2004; Mavrodi et al., 2010; Liu et al., 2015) are also shown in Figure 2. These amino acids residues were also found in PhzF of Pt12, however in different positions.
Figure 2: Alignment between the putative PhzF protein of Pt12 isolate and PhzF proteins from others Pseudomonas bacteria. P. fluorescens 2-79 (AAC18905.1), P. aeruginosa PAO1 (AAC64492.1), P. putida DOTT1E (AFO48928.1), HB3267 (AGA74580.1) and S16 (AEJ14259.1). E45, G73, H74, D208, G212 and S213, indicated by arrows, are conserved amino acids with crucial importance for catalytic activity of PhzF proteins of P. fluorescens and P. aeruginosa.
Molecular modeling of PhzF from th e Pt12 isolate
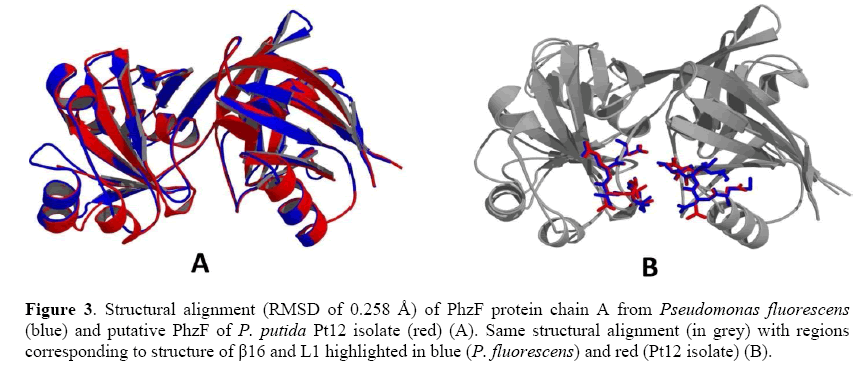
In this study, the template chosen was the PhzF of P. fluorescens (PDB CODE 1XUA), which showed 29% identity, 41% similarity and an alignment score of 71.63 bits with the putative PhzF protein of the Pt12 isolate. This structure was solved by x-ray diffraction with 1.9 Å resolution. The primary sequence of the template presented 596 residues, with each chain containing 298 amino acid residues. The best model validation is presented in Table 2, and the superposition of the template and target main chains based on the Cα, had an RMSD of 0.258 Å (Figure 3).
| M odel | Ramachandran | Verify3D | Qmean | RM SD | Do main |
|---|---|---|---|---|---|
| Pseudomonas putida P zF | 91.09% | 95.33% | -2.50 | 0. 258 | YHI9 (COG0384) |
Table 2: Model identification and validation. Ramachandran values show the residues in favorable regions.
Figure 3: Structural alignment (RMSD of 0.258 Å) of PhzF protein chain A from Pseudomonas fluorescens (blue) and putative PhzF of P. putida Pt12 isolate (red) (A). Same structural alignment (in grey) with regions corresponding to structure of β16 and L1 highlighted in blue (P. fluorescens) and red (Pt12 isolate) (B).
Molecular Dynamics of PhzF of Pt12 and DHHA interaction
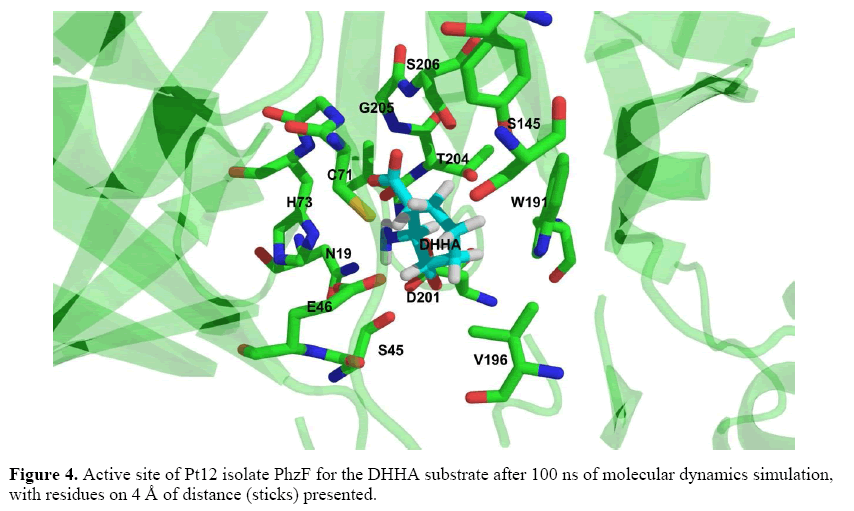
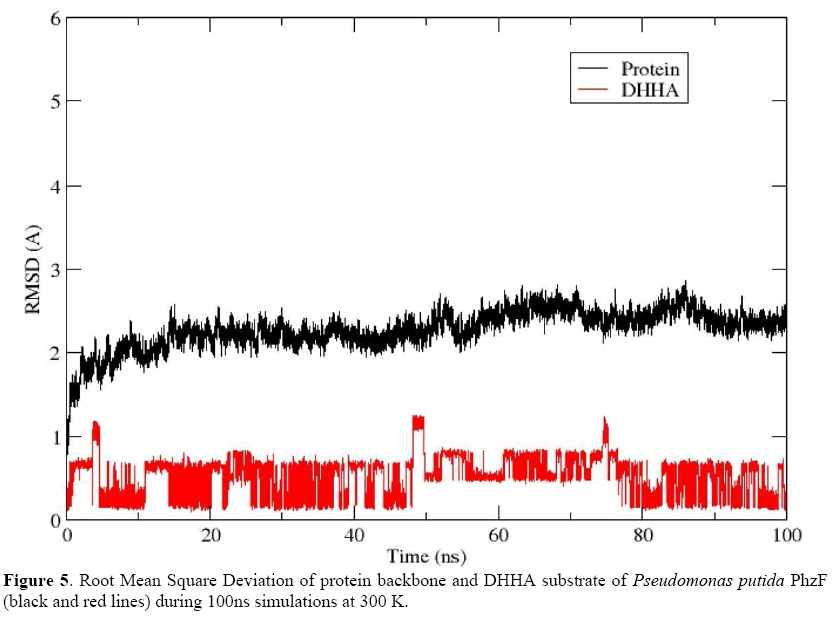
A production run of 100 ns was generated for the Pt12 isolate PhzF protein complexed with DHHA ligand and the ligand remained near the initial binding site (Figure 4). The RMSD was ~2 Å for most of the trajectory (Figure 5), thus validating the model generated by comparative modeling. In this case, the binding free energy calculations were performed based on the last 5,000 frames and MM-GBSA and MM-PBSA results were - 13.84 kcal/mol and -7.43 kcal/mol, respectively. These results indicate a spontaneous association between the model and the DHHA substrate, through interaction with the E46, D201, T204, G205 and S206, among other amino acid residues (Figure 4).
Discussion
In recent years, the use of Pseudomonas bacteria has emerged as a powerful tool in biological control of pathogens of many crops, taking into account mainly their advantages in comparison with chemical products. In this regard, our previous studies identified the Pt12 isolate as a P. putida associated with roots of the Amazonian P. tuberculatum, with potential to control F. solani f. sp. piperis, the causal agent of root rot disease in black pepper (Nascimento et al., 2015).
Here, we aimed to characterize the P. putida Pt12 isolate by detection of sequences related to antifungal metabolites biosynthesis, such as phenazine, DAPG, pyrrolnitrin, pyoluteorin and HCN. Among primers for metabolites tested in the Pt12 isolate, our results showed amplification of a PhzF sequence potentially related to phenazine biosynthesis.
Phenazines are antibiotic metabolites with potent activity against bacteria and fungi. The phenazine biosynthetic pathway in Pseudomonas bacteria requires an operon constituted of at least seven genes, PhzABCDEFG, with the PhzF gene encoding the PhzF protein responsible for DHHA condensation to PCA (Mavrodi et al., 1998, 2001a; Bilal et al., 2017). PhzF is a symmetric homodimer of two subunits, with 278 amino acids per subunit, containing one active site with the conserved catalytically critical residues E45, H74 and D208, according to structural analyses (Parsons et al., 2004; Blankenfeldt et al., 2004; Mavrodi et al., 2010; Liu et al., 2015). It is known that PhzF proteins have structural similarities to other epimerases, but with differences in their catalytic sites. Among them, the diaminopimelate epimerase (DAP), responsible for epimerization of L,L-diaminopimelate to L,D-meso-diaminopimelate through catalytic activity attributed to two conserved cysteines: basic C73 and acidic C217. Members of the YddE/YHI9 family have features common to both PhzF and DAP, including a conserved basic cysteine (Blankenfeldt et al., 2004).
Although the operon for the phenazine biosynthetic pathway has been well characterized in P. fluorescens, P. chlororaphis and P. aeruginosa (Bilal et al., 2017), in other Pseudomonas bacteria, including P. putida, it remains to be elucidated. However, there are many sequences containing putative PhzF domains available in the GenBank Database. In this context, we isolated a 789-bp sequence coding for a 262 aa PhzF-like protein with high identity with PhzF proteins from P. putida (WP_046785327.1) and P. plecoglossicida (WP_041505738.1). According to our analysis, the PhzF of the Pt12 isolatecontains putative conserved domains for two main protein families: COG0384 (YHI9 Predicted epimerase YddE/YHI9) and pfam02567 (PhzC-PhzF, Phenazine biosynthesis-like protein), both belonging to the cl27516 PhzC-PhzF Superfamily (Marchler-Bauer et al., 2017).
Due to the lack of significant identity of the Pt12 isolate PhzF with PhzF proteins that have been functionally evaluated, we compared this sequence with structurally well-characterized PhzF proteins of P. fluorescens (AAC18905) and P. aeruginosa (AAC64492.1), in order to search for conserved amino acid residues that are potentially involved in its function. When we aligned these sequences, we found in Pt12 PhzF the amino acids E46, H73 and D201, which could correspond to the conserved E45, H74 and D208, critical to the catalytic activity of PhzF proteins (Parsons et al., 2004; Blankenfeldt et al., 2004; Mavrodi et al., 2010; Liu et al., 2015). The G72, G205 and S206 were other amino acid residues potentially involved in the active site of Pt12 isolate PhzF. According to Liu et al. (2015), the conserved E45 at the active site of PhzF acts as a general base/acid catalyst transferring a proton from C3 to C1 of DHHA, while G73, H74, D208, G212 and S213 form hydrogen bonds with the substrate to stabilize proton transfer. As shown in Figure 2, the E45/E46 is part of the NLSE sequence, which seems to be a conserved motif found in all PhzF sequences used in our alignment, including other putative PhzF sequences of P. putida.
Collectively, the conserved amino acids found in Pt12 isolate PhzF indicated the possibility of an active site for the DHHA ligand within this protein. Then, we predicted the interaction between this protein and the DHHA using molecular modeling and molecular dynamic simulations (MDS). As reported by Karplus and McCammo (2002), molecular dynamics simulations have been used with success in studies of structure and function of biomolecules. In addition, such computational tools are useful for the elucidation of the PhzF catalysis mechanism, as shown by Liu et al. (2015).
For molecular modeling of Pt12 PhzF, we used the P. fluorescens PhzF as a template, containing 8 α-helixes (α1-α8), 16 β-sheets (β1-β16), and about 20 loop regions (L1-L20), as shown by Liu et al. (2015). Molecular dynamics simulations revealed a spontaneous and stable association between the Pt12 PhzF model and the DHHA substrate, with a RMSD of ~2 Å and binding free energy values of -13.84 kcal/mol and -7.43 kcal/mol for MM-GBSA and MM-PBSA, respectively. Our RMSD value is in accordance with studies reported by Liu et al. (2015), who found RMSD values range from 1.74 to 1.84 for PhzF-DHHA complexes. Besides, most of the amino acid residues of Pt12 PhzF interacting with DHHA were those identified in our alignment and have been described as related to the active site of the PhzF template. For example, E46 could participate by proton transfer, as described for E45 in P. fluorescens PhzF (Liu et al., 2015). Likewise, D201, T204, G205 and S206 were also found in the Pt12 PhzF and DHHA interaction, where they could play similar roles to D208, T211, G212 and S213 in P. fluorescens PhzF.
Regarding D208, this conserved amino acid residue can form a hydrogen-bonded network with N18, S44, A210, T211, and the ligand amino group at the active site (Blankenfeldt et al., 2004). Then, it is possible that D201 acts together with N19, S45 and T204 in the formation of a hydrogen-bonded network required for the conversion of DHHA to PCA by the Pt12 isolate PhzF.
Besides the biochemical roles of each of the conserved amino acid residues in the catalytic activity of P. fluorescens PhzF, according to Liu et al. (2015) the structure of β16 (residues Y203-G204-V205-V206-E207-D208) and L1 (residues M41-N42-L43-S44-E45) could contribute to the formation of a passage leading to the enzyme active site. Interestingly, most of these amino acids are also found in Pt12 PhzF (V196-G197-V198-N199-E200-D201 and N42-N43-L44-S45-E46), and as shown in Figure 3, the superposition of these two proteins revealed an RMSD of 0.258 Å and structural similarities between them.
In contrast to the above mentioned amino acid residues, our MDS also revealed C71, S145, W191 and V196, which, as far as we know, are amino acids that are not described in active sites of other PhzF proteins. Among them, cysteine residues have been found in catalytic sites of other epimerases, such as the YddE from Escherichia coli, which contains a conserved basic cysteine from DAP and also some catalyticresidues of PhzF. However, this protein does not use DHHA as a substrate (Blankenfeldt et al., 2004). On the other hand, our MDS results showed Pt12 PhzF could stably interact with DHHA in phenazine biosynthesis. Besides the conserved amino acids described in active site of well-characterized PhzF proteins, the interaction of Pt12 PhzF with DHHA could also involve other amino acids, including C71, which needs to be functionally investigated.
In addition to the detection of bacterial genes related to phenazine biosynthesis, other studies have confirmed P. putida strains as producers of phenazine and its derivatives. For instance, El-Sayed et al. (2008) reported P. putida strains producing between 35.8 to 40.0 ug/ml of PCA with potential to control Alternaria solani. Likewise, Liu et al. (2006) evaluated 48 Pseudomonas spp. from the green pepper rhizosphere and found isolates closely related to P. putida producing phenazine (PCA). Recently, Kennedy et al. (2015) identified the 5-methyl phenazine-1-carboxylic acid betaine as a potent antimicrobial and anticancer metabolite produced by P. putida. Based on data from the literature and our results presented here, we suggest that Pt12 PhzF could be a functional protein in phenazine production by the P. putida Pt12 isolate; however, additional studies will be needed to confirm its role.
Conclusion
Here, we report for the first time the detection of sequences related to antifungal metabolite biosynthesis in the P. putida Pt12 isolate, with the identification of a putative PhzF protein able to interact stably with DHHA according to MDS, contributing to the molecular characterization of this rhizobacterium with potential to control root rot disease of black pepper in the Amazon region.
Acknowledgment
The authors thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação Amazônia de Amparo a Estudos e Pesquisas do Pará (FAPESPA), Universidade do Estado do Pará (UEPA), and the Universidade Federal do Pará (UFPA), Brazil.
Conflicts of interest
The authors declare no conflicts of interest.
About the Authors
Corresponding Author
C.R.B. Souza
Instituto de Ciências Biológicas, Universidade Federal do Pará, Belém, PA, Brasil
- Email:
- bsouza@ufpa.br
References
- Albuquerque FC, Duarte MLR, Benchimol RL, Endo T (2001). Resistência de piperáceas nativas da Amazônia à infecção causada por Nectria haematococca f. sp. piperis. Acta Amaz. 31: 341-348.
- Albuquerque FC and Ferraz S (1976). Características morfológicas e fisiológicas de Nectria haematococca f. sp. piperis e sua patogenicidade à pimenta do reino (Piper nigrum L.). Experientia 22: 133-151.
- Allaire M (2005). Diversite fonctionnelle des Pseudomonas producteurs d’antibiotiques dans les rhizospheres de coniferes en pepinieres et en milieu naturel. Master Dissertation, Laval Universitz, France.
- Altschul SF, Madden TL, Schäffer AA, Zhang J, et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402.
- Beneduzi A, Ambrosini A, Passaglia LMP (2012). Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 35: 1044–1051.
- Benkert P, Tosatto SCE, Schomburg D (2008). QMEAN: A comprehensive scoring function for model quality assessment. Proteins 71: 261–277.
- Bilal M, Guo S, Iqbal HMN, Hu H, et al. (2017). Engineering Pseudomonas for phenazine biosynthesis, regulation, and biotechnological applications: a review. World J. Microbiol. Biotechnol. 33: 191.
- Blankenfeldt W, Kuzin AP, Skarina T, Korniyenko Y, et al. (2004). Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens. Proc. Natl. Acad. Sci. U S A. 101: 16431-16436.
- Cornea CP, Ciuca M, Voaides C, Popa G, et al. (2007). Molecular detection of the genes involved in the antifungal properties of some Pseudomonas spp. Bulletin UASVM – CN. 64.
- Davis IW, Leaver-Fay A, Chen VB, Block JN, et al. (2007). MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35: W375–W383.
- El-Sayed W, Abd El-Megeed M, Abd El-Razik AB, Soliman KH, et al. (2008). Isolation and identification of phenazine-1-carboxylic acid from different Pseudomonas isolates and its biological activity against Alternaria solani. Res. J. Agric. Biol. Sci. 4: 892-901.
- Genheden S, Ryde U (2015). The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 10: 449–461.
- Glick BR (2012). Plant growth-promoting bacteria: mechanisms and applications. Scientifica 2012: 963401.
- Hall TA (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41: 95–98.
- Hassan MN, Afghan S, Hafeez FY (2011). Biological control of red rot in sugarcane by native pyoluteorin-producing Pseudomonas putida strain NH-50 under field conditions and its potential modes of action. Pest Manag. Sci. 67:1147–1154.
- IBGE (Instituto Brasileiro de Geografia e Estatística) (2016). Levantamento sistemático da produção agrícola: pesquisa mensal de previsão e acompanhamento das safras agrícolas no ano civil. IBGE, Rio de Janeiro.
- Karplus M and McCammon JA (2002). Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 9: 646–652. Kennedy RK, Naik PR, Veena V, Lakshmi BS, et al. (2015). 5-Methyl phenazine-1-carboxylic acid: a novel bioactive metabolite by a rhizosphere soil bacterium that exhibits potent antimicrobial and anticancer activities. Chem. Biol.Interact. 231: 71-82.
- Keshavarz-Tohid V, Taheri P, Muller D, Prigent-Combaret C, et al. (2017). Phylogenetic diversity and antagonistic traits of root and rhizosphere pseudomonads of bean from Iran for controlling Rhizoctonia solani. Res. Microbiol. 168: 760–772.
- Liu H, Dong D, Peng H, Zhang X, et al. (2006). Genetic diversity of phenazine- and pyoluteorin-producing pseudomonads isolated from green pepper rhizosphere. Arch. Microbiol. 185: 91-98.
- Liu F, Zhao YL, Wang X, Hu H, et al. (2015). Elucidation of enzymatic mechanism of phenazine biosynthetic protein PhzF using QM/MM and MD simulations. PLoS ONE 10: e0139081.
- Lüthy R, Bowie JU, Eisenberg D (1992). Assessment of protein models with three-dimensional profiles. Nature 356: 83– 85.
- Maier JA, Martinez C, Kasavajhala K, Wickstrom L, et al. (2015). ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11: 3696–3713.
- Marchler-Bauer A, Bo Y, Han L, He J, et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45: D200-D203.
- Mavrodi DV, Ksenzenko VN, Bonsall RF, Cook RJ, et al. (1998). Seven-gene locus for synthesis of phenazine-1-carboxylic acid by Pseudomonas fluorescens 2-79. J. Bacteriol. 180: 2541–2548.
- Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, et al. (2001a). Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 183: 6454 -6465.
- Mavrodi DV, McSpadden Gardener BB, Mavrodi DV, Bonsall RF, et al. (2001b). Genetic diversity of phlD from 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. Phytopathology 91: 35-43.
- Mavrodi DV, Peever TL, Mavrodi OV, Parejko JA, et al. (2010). Diversity and evolution of the phenazine biosynthesis pathway. Appl. Environ. MicroBiol. 76: 866–879.
- McSpadden Gardener BB, Mavrodi DV, Thomashow LS, Weller DM (2001). A rapid polymerase chain reaction-based assay characterizing rhizosphere populations of 2,4-diacetylphloroglucinol producing bacteria. Phytopathology 91: 44–54.
- Nascimento SB, Lima AM, Borges BN, de Souza CRB (2015). Endophytic bacteria from Piper tuberculatum Jacq.: isolation, molecular characterization, and in vitro screening for the control of Fusarium solani f. sp piperis, the causal agent of root rot disease in black pepper (Piper nigrum L.). Genet. Mol. Res. 14: 7567-7577.
- Parsons JF, Song F, Parsons L, Calabrese K, et al. (2004). Structure and function of the phenazine biosynthesis protein PhzF from Pseudomonas fluorescens 2-79. Biochemistry 43: 12427–12435.
- Pei J, Tang M, Grishin NV (2008). PROMALS3D web server for accurate multiple protein sequence and structure alignments. Nucleic Acids Res. 36: W30–W34.
- Puopolo G, Masi M, Raio A, Andolfi A, et al. (2013). Insights on the susceptibility of plant pathogenic fungi to phenazine-1-carboxylic acid and its chemical derivatives. Nat. Prod. Res. 27: 956-966.
- Raaijmakers JM, Weller DM, Thomashow LS (1997). Frequency of antibiotic-producing Pseudomonas spp. in natural environments. Appl. Environ. Microbiol. 63: 881–887.
- Ramette A, Frapolli M, Défago G, Moënne-Loccoz Y (2003). Phylogeny of HCN synthase-encoding hcnBC genes in biocontrol fluorescent pseudomonads and its relationship with host plant species and HCN synthesis ability. Mol. Plant Microbe In. 16: 525- 535.
- Sarhan EAD and Shehata HS (2014). Potential plant growth-promoting activity of Pseudomonas spp. and Bacillus spp. as biocontrol agents against damping-off in alfalfa. Plant Pathology J. 13: 8-17.
- Vanitha S and Ramjegathesh R (2014). Bio control potential of Pseudomonas fluorescens against coleus root rot disease. J. Plant Pathol. Microb. 5: 216.
- Verhagen BW, Trotel-Aziz P, Couderchet M, Höfte M, et al. (2010). Pseudomonas spp.-induced systemic resistance to Botrytis cinerea is associated with induction and priming of defence responses in grapevine. J. Exp. Bot. 61: 249-260.
- Webb B and Sali A (2016). Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 54: 5.6.1-5.6.37.
- Zhang Y, Chen P, Ye G, Lin H, et al. (2018). Complete genome sequence of Pseudomonas parafulva PRS09-11288, a biocontrol strain produces the antibiotic phenazine-1-carboxylic acid. Curr. Microbiol. https://doi.org/10.1007/s00284-018-1441-0.
Keywords:
Download:
Full PDF- Share This