Evaluation of genetic variability and association of traits in Kabuli chickpea (Cicer arietinum L.) genotypes in East Gojjam Zone, Northwestern, Ethiopia
Received: September 02, 2024
Accepted: September 04, 2024
Published: September 30, 2024
Genet.Mol.Res. 23(3):
Keywords
Correlation; Heritability; Path coefficient; Genetic advance; Genetic parameter
Introduction
Chickpea (Cicer arietinum L.) is grown all over the world in about 57 (fifty-seven) countries including Ethiopia. India is the leading producer [1]. Chickpea is also the third most important annual cool season food grain legumes in the world after common bean (Phaseolus vulgaris L.) and field pea [2,3]. This crop has a small genome size of 740 Mbp and is self-pollinated. Further, chromosomal constitution reveals chickpea as a diploid species with 2n=2x=16chromosomes [4]. The crop is grown under wide agro ecological conditions in midlands with an altitude of 1500 2300 mASL and rainfall of 7001 up to 300mm in Ethiopia [5]. It is commonly cultivated in vertisol type using residual moisture offers to the farmers for double cropping. Chickpeas do have multiple roles in the farming systems of many developing countries including Ethiopia ranging from human food, animal feed, export commodities and provide environmental services [1,6,7]. Nutritionally, chickpea is the cheapest and most radially available source of protein, fat, carbohydrate, fiber and minerals and vitamins [8]. Ethiopia is the sixth largest chickpea producing country sharing 3.15% of the world's total production, first in Africa shares about (63%) of the total production, 4.5% of the global chickpea market and more than 60% of Africa’s market [9]. The crop is the third food grain legume in Ethiopia in terms of area and production [10]. Its productivity was (2.18), next to soya bean (2.49) and fababean (2.22) tha-1. Chickpea cultivation in Amhara Region was 0.1 million hectares in production of 0.2 million tons with productivity of 1.96 tha-1. East Gojjam Zone accounts 0.008 million ha of cultivated area and 0.02 million tons of production with productivity of 2.3 tha-1. Awabel district covers 200 hectares of land [11].
However, the national average seed yield in Ethiopia is low, 2.18 tons ha-1 far below its genetic potential yield of 2.9 tons ha-1 [12,13], because the production of the crop nationally constrained by many factors such as usage of inappropriate improved varieties, use of inherent low productive farmers varieties, biotic and abiotic factors [6,14,15]. To alleviate some of these problems, many chickpea breeding efforts have been done since its beginning in the 1970’s in Ethiopia to improve production and productivity. In its age of breeding approximately, 29 super performing varieties were released in both types; most of these were developed from the breeding material supplied by ICRISAT and ICARDA [6]. Nevertheless, most of the developed verities were Desi type. For a successful improvement of kabuli type chickpea fairly, the presence of genetic variability plays vital role. The selection of superior genotypes depends on the variability of genotypes [10]. The success of improvement in crop breeding depends upon the magnitude of genetic variability available in breeding material and the extent of heritability of desirable traits [16-18].
Yield is a complex quantitative trait that is dependent on associations and direct and indirect effects of many yield contributing traits [19]. Thus, it helps in the determination of the selection criteria for simultaneous improvement of various traits along with economic yield [20]. Therefore, correlation in combination with path coefficient analysis will be an important tool to realize the association and quantify the direct and indirect influence of one trait upon another [21]. Understanding the genetic source of yield and yield traits in addition to genetic variation and relationships between genotypes is vital to exploit the existing genetic variability and its potential use in breeding programs [22]. The demand for a variety with high yield over a range of production environments is very high among chickpea growers. Hence, genetic improvement to develop varieties with high yield potential, wider adaptability and resistance/tolerance to biotic and abiotic stresses, with acceptable end use quality, is the most viable and environment friendly option to sustainably increase chickpea yield [6].
Such improvement of crops requires the creation and introduction of genetic variability, in breeding coupled with selection, and extensive evaluation of breeding materials at multiple locations/ seasons to identify adapted and stable genotypes with desirable agronomic traits [23,24]. Still, variability of the quantitative traits of crops is the great interest to researchers to develop a new variety [25]. Previous studies were carried out on genetic variability of chickpeas with the help of genetic parameters, correlation coefficient and path analysis to determine the important traits and improvement of various traits along with economic yield for a different location in Ethiopia. But, little information is available in northwestern parts of the country [11,26-28]. Besides, the availability of genetically improved Kabuli chickpea varieties has little enough and limited efforts have been made to improve the crop through the study of Kabuli chickpea genetic variability for major traits such as yield [29]. Even though different research efforts have been done for the development of improved varieties across the country in Ethiopia, there is a limitation of Kabuli chickpea variety development work and popularization efforts from the national research system in East Gojjam Zone for selecting well-adapted and high yielding genotypes [30]. Therefore, it is important to estimate the magnitude of genetic variability for yield, yield related traits of Kabuli chickpea genotypes, association among traits, and determine the direct and indirect effect of yield related traits on seed yield.
Materials and Methods
Description of the study area
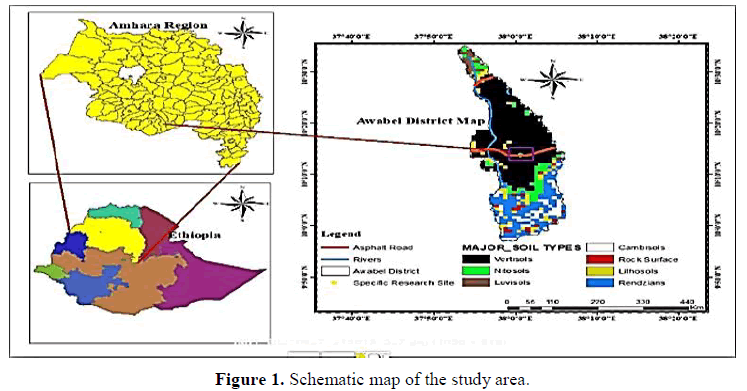
The experiment was conducted in East Gojam Zone, Northwestern Ethiopia during two seasons the 2019/2020 and 2020/2021 main cropping season. The study area was found at a distance of 258 km northwest of Addis Ababa. Geographically, the site lies at 10° 29' latitude north and 37° 44' longitude east with an altitude of 2104 mAMSL. It receives a mean annual rainfall of about 1090 mm. The mean minimum and maximum annual temperatures are 15°C and 24°C, respectively. The soil type of the site is predominantly black vertisols with a pH value of 6.45 (Figure 1).
Experimental materials
Thirty-six Kabuli chickpea genotypes including two standard check varieties (Arerti and Habru) were included in the study. These experimental materials were obtained from the highland pulse research program, Debre Zeit Agricultural Research Center (DZARC) (Table 1).
| Code | Genotype pedigree | Code | Genotype pedigree | Code | Genotype pedigree |
|---|---|---|---|---|---|
| G-1 | FLIP-93-93C | G-13 | Arerti | G-25 | FLIP-12-343C |
| G-2 | FLIP-12-53C | G-14 | FLIP-12-60C | G-26 | FLIP-12-311C |
| G-3 | FLIP-12-110C | G-15 | FLIP-12-57C | G-27 | FLIP-12-176C |
| G-4 | FLIP-12-37C | G-16 | FLIP-12-86C | G-28 | FLIP-12-40C |
| G-5 | Habru | G-17 | FLIP-12-342C | G-29 | FLIP-12-01C |
| G-6 | FLIP-12-198C | G-18 | FLIP-12-263C | G-30 | FLIP-12-331C |
| G-7 | FLIP-12-107C | G-19 | FLIP-82-150C | G-31 | FLIP-12-265C |
| G-8 | FLIP-12-287C | G-20 | FLIP-88-85C | G-32 | FLIP-12-55C |
| G-9 | FLIP-12-06C | G-21 | FLIP-12-108C | G-33 | FLIP-12-197C |
| G-10 | FLIP-12-18C | G-22 | FLIP-12-322C | G-34 | FLIP-12-210C |
| G-11 | FLIP-12-79C | G-23 | FLIP-12-310C | G-35 | FLIP-12-75C |
| G-12 | FLIP-12-61C | G-24 | FLIP-12-109C | G-36 | FLIP-12-192 |
Table 1. List of Kabuli chickpea genotypes used in the experiment.
Experimental design, procedures and trial management
The experiment was carried out in a 6 × 6 simple lattice design during the 2019/2020 and 2020/2021 two cropping seasons. The total plot size was 2.25 m2 (1.5 m length × 1.5 m width) with a net plot size of 1.35 m2. Each genotype was planted with a spacing of 30 cm between rows and 10 cm between plants. NPS fertilizers were applied as recommended (121 kg ha-1) and all other agronomic practices were done as per the recommendation for chickpea production.
Data collection
The data were collected from the net plot area at randomly selected and tagged five individual plants by adopting [31,32]. The data were collected on plot basis are days to 50% flowering (DF), days to 90% physiological maturity (DM), Pod Filling Period (PFP), Hundred Seed Weight (HSW) (g), Biomass Yield (BY), Seed Yield (SY) and Harvest Index (HI). While the data collected on plant basis were Plant Height (PH) (cm), Number of Primary Branches (NBP), Number of Secondary Branches (NSB), Number of Pods per Plant (NPP), Number of Seeds Per Pod (NSP) and Number of Seeds Per Plant (NSPP).
Data analysis
Analysis of variance: All measured traits were subjected to Analysis of Variance (ANOVA) using PROC GLM of SAS statistical software version 9.4 to assess the difference among the tested genotypes. Comparison of treatment means were tested using Least Significant Difference test (LSD) at 5% of probability level [33]. The relationship between traits with yield and yield components were correlated using the Proc-Corr Pearson’s correlation procedures.
Estimation of genetic parameters
Estimation of variances components: The phenotypic variance, genotypic variance and coefficients of variations were estimated by the procedure suggested by Burton GW et al. [34].
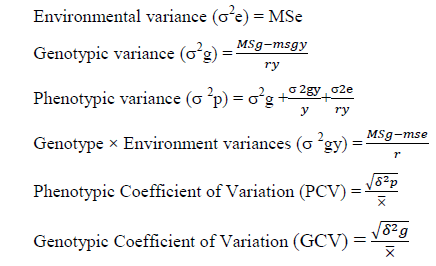
Where, ̅=grand mean of a character, r=replication, msgy=genotypic x year mean square, MSg=genotypic mean square and MSe=mean square of error
PCV and GCV values ranging from >20% are regarded as high, values 10% to 20% as a medium,<10% are considered as suggested by Deshmukh S et al. [35].
Heritability in a broad sense: Heritability (H2) is expressed as a percentage of the ratio of the genotypic variance (σ2 g) to the phenotypic variance (σ2p) as follows [36]:

Where,
H2=heritability in a broad sense, σ2p=phenotypic variance, σ2g=Genotypic variance. According to Singh, heritability values were categorized as high greater than 60 moderate 40-59% and less than 40% is engaged as low.
Genetic Advance (GA): According to Allard [36], selection intensity (K) at 5% was computed as follows:
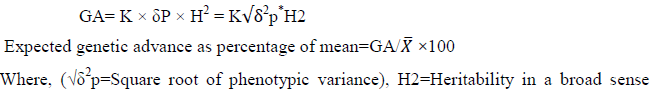
x=grand populations mean for the trait under considerations, K=is a constant value at Standardized selection differential (where k=2.06 at 5% selection intensity) σp=phenotypic variance
Genetic advance as a percent of the mean range was categorized as low (<10%), moderate (10 up to 20%) and high (>20%) as suggested by Johnson P, et al. [37].
Association of traits
Estimation of phenotypic and genotypic associations: Phenotypic and genotypic correlations were estimated using the formula suggested by Miller P, et al. [38].
Path coefficient analysis: Path coefficient analysis was computed by using the suggested formula by Wright [39] and worked out by Dewey and Lu [21] using the phenotypic correlations to determine the direct and indirect effect of yield components on seed yield [20,38]. The residual effect (U) is the unexplained variation of the trait that is not accounted for by the path coefficient and is calculated using the formula of Dewey DR, et al., Singh RK, et al. [21,40].
Results and Discussion
Analysis of variance
The homogeneity test of variances over two years showed the f-ratio was <3 for all traits. ANOVA has shown a highly significant difference among traits of all studied genotypes (Table 2). ANOVA has also shown significant difference among traits over years, except the trait plant height and 100-seed weight. Combined ANOVA indicated that the interaction effects of genotype and year were significant for most traits such number of pods per plant, number of seeds per plant, number of seeds per pod, biomass yield, hundred seed weight and seed yield. This indicates the existence of high genetic variability that endowed the breeder for future improvement. Our result was corroborated with the results of many agronomic traits and in chickpea genetic variability and association study [2,40-43,27].
| Mean squares | ||||||||
|---|---|---|---|---|---|---|---|---|
| Traits | Replication | Block | Genotype (G) | Year (Y) | G*Y | Error | CV | R2 |
| DF=1 | DF=5 | DF= 35 | DF=1 | DF=35 | DF=143 | |||
| DF | 44.4 | 6.87 | 133.77** | 250.69** | 11.22 | 12.34 | 6.13 | 0.87 |
| PFP | 12.25 | 106.78** | 194.24** | 8130.03** | 18.32 | 27.33 | 7.06 | 0.89 |
| DM | 5.06 | 144.37** | 340.22** | 15149.5** | 11.52 | 35.57 | 4.58 | 0.92 |
| PH | 0.095 | 59.85** | 63.50** | 0.0034 | 63.92** | 18.62 | 9.06 | 0.79 |
| NPB | 0.33 | 0.112 | 7.21** | 5.56** | 0.95** | 0.21 | 11.56 | 0.95 |
| NSB | 47.61** | 0.49 | 3.19* | 26.01** | 4.25** | 2 | 20.95 | 0.72 |
| NPP | 13.93 | 39.68 | 554.49** | 30531.73** | 310.06** | 19.8 | 9.23 | 0.97 |
| NSP | 0.0015 | 0.0049 | 0.046** | 0.067** | 0.0126** | 0.0041 | 5.55 | 0.88 |
| NSPP | 8.507 | 75.68* | 521.66** | 2789.6** | 83.23** | 27.2 | 15.04 | 0.93 |
| HSW | 55 | 76.33 | 146.69** | 8.5 | 81.88** | 34.19 | 18.35 | 0.78 |
| SY | 465487.8 | 171230.7 | 4553790.5** | 3989873.1** | 335634.0* | 209955.6 | 15.57 | 0.93 |
| BY | 1544736 | 4483217 | 19611924.7** | 29606376.3** | 10414631.4** | 2829656 | 21.13 | 0.86 |
| HI | 0.037 | 0.023 | 0.032** | 0.00075** | 0.035 | 0.01088 | 26.49 | 0.78 |
Note:*: significant at (p ≤ 0.05) and **: Highly significant (p ≤ 0.01) probability level. CV: Coefficient of variation;
DF: Days to 50% Flowering; DM: Days to 90% Maturity; PFP: Pod Filling Period; PH: Plant Height;
NPB: Number of Primary Branches; NSB:Number of Secondary Branches;
NPP: Number of Pods Per Plant; NSPP: Number of Seeds per Plant;
NSP: Number of Seeds per Pod; BY: Biomass Yield;
HSW: Hundred Seed Weight; SY: Seed Yield and HI: Harvest Index.
Table 2. Combined analysis of variance for 13 traits of tested genotypes over years.
Mean and range values
Seed yield ranged from 929.5-5530.4 kgha-1 with a mean value of 2948.69 kgha-1. The highest seed yield was recorded from genotype FLIP-93-93C (5530.4 kgha-1) followed by FLIP-12-198C (5033.4 kgha-1) and FLIP-12-343C (4406.8 kgha-1). Genotype FLIP-93-93C it could be most promising for seed yield improvement of the Kabuli chickpea. The highest biomass yield was recorded for FLIP-12-37C and FLIP-12-57C (133001 kgha-1). Chickpea straw has important role for animal feed and soil improvement. Hence, genotype FLIP-12-37C and FLIP-12-57C will be considered in biological yield improvement of Kabuli chickpea to address farmers need and improve animal production fairly. Similar results were noted in the previous study on Kabuli chickpea genotypes (Tables 3 and 4) [27,42,44,45].
| Traits | |||||||
|---|---|---|---|---|---|---|---|
| Gen | DF | PFP | DM | PH | NPB | NSB | NPP |
| 1 | 60b-e | 80.5a-f | 140.25bc | 50.62b-g | 5.47a-e | 7.37a-f | 70.75a |
| 2 | 53f-j | 66.5lmn | 118.25no | 51.92bcd | 4.6e-g | 6.15c-i | 45.05j-n |
| 3 | 53f-j | 73j-l | 124.75i-n | 49.12c-g | 5e-g | 6.57b- | 53.77efg |
| 4 | 44.75k | 66.75lmn | 110.25lmn | 49.17c-g | 3.55hij | 7.07a-h | 47.52g-m |
| 5 | 50.75ij | 74.25d-k | 123.75j-n | 42.47ijk | 2.2pq | 5.87d-i | 34.57qrs |
| 6 | 48.25jk | 70.5i-l | 118.5mno | 46.97c-j | 5.77abc | 8.42ab | 57.77de |
| 7 | 54.75f-i | 81.25a-e | 135.25c-j | 48.25c-i | 5.1d-f | 7.42a-f | 51.47f-i |
| 8 | 49.75j | 73.25f-l | 121.75lmn | 52bcd | 3.37h-k | 6.57b-h | 50.77f-i |
| 9 | 57.75c-f | 69.75j-m | 126.25i-n | 46.85c-j | 4.37g | 7.2a-h | 43.6l-p |
| 10 | 55f-i | 72h-l | 125.75i-n | 47.07c-j | 2.02pq | 5.55f-i | 50.6f-k |
| 11 | 64.75ab | 62.75mno | 126.25i-n | 46.6c-j | 6a | 8abc | 65.82abc |
| 12 | 55.75e-h | 74.75 d-k | 129f-l | 45.82e-k | 1.77q | 6.1c-i | 28.9s |
| 13 | 53f-j | 60.5no | 112.25o | 42.82h-k | 2.4l-q | 6.32c-h | 33.82rs |
| 14 | 56.75e-h | 73.75e-l | 130.25e-k | 46.25d-k | 2.82k-o | 7.12a-h | 32.65rs |
| 15 | 66.5a | 83.5ab | 148.75a | 50.27b-g | 3i-l | 7.45a-f | 44.42k-n |
| 16 | 50.5ij | 78.25a-h | 127.5g-l | 42.5ijk | 5.92ab | 7.27a-h | 68.47abc |
| 17 | 66.25a | 83.75ab | 148.75a | 44.57g-k | 2.32n-q | 5.35ghi | 42.82m-p |
| 18 | 65.75a | 81.25a-d | 145.75ab | 49.55b-g | 1.75q | 5.7e-i | 30.75s |
| 19 | 66a | 89o | 123.75j-n | 44.9f-k | 5.32b-e | 6.725b-h | 52.4e-h |
| 20 | 57.5d-g | 82.5abc | 138.75bcd | 49.9b-g | 5.7a-d | 7.32a-g | 50.57f-k |
| 21 | 63.5ab | 77b-j | 140.25bc | 50.95b-d | 3.15h-l | 6.725b-h | 33.47rs |
| 22 | 62a-d | 61.25no | 122k-n | 55.62ab | 3.7h | 5.3hi | 53.77efg |
| 23 | 57e-h | 76.5b-j | 132.25c-i | 45.45e-k | 5.75a-d | 8abc | 62.4cd |
| 24 | 62a-d | 75.5c-k | 136.25c-f | 46.47d-j | 6a | 7.2a-h | 68.07bc |
| 25 | 52.75g-j | 69.75j-m | 121.2lmn | 44.82g-k | 4.97e-g | 6.72b-h | 45.27i-n |
| 26 | 60.25b-e | 77.25b-i | 136.25c-f | 52.62bc | 5.42a-e | 8.8a | 63.87bcd |
| 27 | 52.75hij | 77.75a-i | 130.25e-k | 44.55g-k | 3.17h-l | 6.175c-i | 38.05o-r |
| 28 | 50.25ij | 83.25ab | 132.25c-i | 51.27b-e | 4.97e-g | 6.95a-h | 42.1m-p |
| 29 | 60b-e | 66.5lmn | 125.25i-n | 60.875a | 2.6l-p | 4.275i | 37.15pqr |
| 30 | 56.7e-h | 82.75abc | 138.25b-e | 46.85c-j | 2.7l-o | 5.6e-i | 41.9m-p |
| 31 | 64.5ab | 71.5h-l | 134.75c-h | 46.4d-k | 4.875e-g | 7.75a-d | 54.12ef |
| 32 | 52.5hij | 79.5a-g | 130.75d-j | 42.92h-k | 2.9j-n | 6.7b-h | 33.52rs |
| 33 | 56e-h | 72h-l | 126.75h-m | 40.32k | 3.52hij | 6.775b-h | 70.75a |
| 34 | 64.75ab | 72.5g-l | 136c-f | 48.77c-h | 5.22c-f | 6.6b-h | 40.42n-q |
| 35 | 62.5abc | 69klm | 130.25e-k | 42.12jk | 2.8k-o | 6.775b-h | 46.37h-n |
| 36 | 52.25hij | 84.75a | 135.75c-g | 45.55e-k | 3.6hi | 7.6a-e | 49.15f-l |
| Range | 44.75-66.5 | 61.25-89 | 110.25-148.75 | 42.12-60.87 | 1.77-6 | 4.27-8.42 | 28.9-70.75 |
| Mean | 57.2 | 74 | 130.11 | 47.59 | 3.99 | 6.76 | 48.2 |
| LSD | 4.96 | 7.38 | 8.47 | 6.09 | 0.65 | 2 | 6.28 |
Table 3. Mean performance of 36 Kabuli chickpea genotypes for yield components and yield evaluated over years at Awabel district.
| Traits | ||||||
|---|---|---|---|---|---|---|
| Gen | NSP | NSPP | HSW(g) | SY (Kg/ha) | BY (Kg/ha) | HI |
| 1 | 1.31a-d | 50.3b | 26.75i-l | 5530.4a | 13301a | 0.437b-g |
| 2 | 1.14g-k | 33.67d-h | 33.75d-i | 3522.2d-g | 9319d-h | 0.383c-l |
| 3 | 1.1i-l | 41.8c | 43.75ab | 3346.5e-h | 9270d-h | 0.365d-l |
| 4 | 1.22-g | 28.42h-k | 32e-k | 4328c | 11773abc | 0.409b-j |
| 5 | 1.25c-f | 34.2d-h | 30.25f-l | 3817.6cde | 11078a-d | 0.386b-k |
| 6 | 1.4a | 70.47a | 29f-l | 5033.4ab | 11573a-d | 0.433b-g |
| 7 | 1.04lm | 28.7h-k | 30.75f-l | 1661.3opq | 7828f-j | 0.270i-l |
| 8 | 1.035lm | 22.4jk | 35.5b-g | 2437.1j-n | 9704c-f | 0.256kl |
| 9 | 1.03lm | 22k | 24.75jkl | 929.5r | 3581n | 0.268jkl |
| 10 | 1.2e-h | 33.1e-i | 44a | 3611.6def | 7463f-l | 0.502a-d |
| 11 | 1.18f-i | 29.67f-j | 31f-l | 2757h-k | 6830i-l | 0.421b-h |
| 12 | 1.18f-i | 22.45jk | 40a-e | 948.5r | 3760mn | 0.296g-l |
| 13 | 1.14g-k | 49.22b | 27.25g-l | 4328c | 7868f-j | 0.641a |
| 14 | 1.14g-k | 26ijk | 34.5c-h | 3029.4f-j | 6438i-m | 0.521abc |
| 15 | 1.21e-h | 24.4jk | 29.5f-l | 3872cde | 13301a | 0.293g-l |
| 16 | 1.16f-j | 49.2b | 26.75i-l | 3365.6e-h | 7857f-j | 0.435b-g |
| 17 | 1.14g-k | 33.42d-h | 34.5c-h | 3844.8cde | 7460f-l | 0.525abc |
| 18 | 1.06kl | 22.3k | 29.75f-l | 3347.6e-h | 7310g-l | 0.468b-f |
| 19 | 1.12h-l | 22.6jk | 26ijkl | 1361.1pqr | 4332mn | 0.416b-i |
| 20 | 0.96m | 22.6jk | 27.5g-l | 2418.4j-n | 5184lmn | 0.477b-e |
| 21 | 1.16f-j | 28.27h-k | 41.5a-d | 3863.4cde | 10774b-e | 0.36d-l |
| 22 | 1.05klm | 35.8c-f | 24kl | 3366.2e-h | 8399e-j | 0.402b-k |
| 23 | 1.12h-l | 49.575b | 24.5kl | 2625.3j-m | 5397k-n | 0.491b-e |
| 24 | 1.05klm | 25jk | 23l | 2248.5k-o | 6790i-l | 0.389b-k |
| 25 | 1.32abc | 64.4a | 24kl | 4406.8bc | 8289f-j | 0.533ab |
| 26 | 1.12h-l | 24.8jk | 35c-h | 2010mno | 5342k-n | 0.41b-j |
| 27 | 1.16f-j | 40cde | 30f-l | 2263.2k-o | 7855f-j | 0.357d-l |
| 28 | 1.18f-i | 37c-f | 42.75abc | 2698.6i-l | 8510e-j | 0.321f-l |
| 29 | 1.4a | 38.85cde | 35.25b-g | 3558.6d-g | 7533f-l | 0.48b-e |
| 30 | 1.03lm | 23.55jk | 33.75d-i | 1928.5nop | 7025h-l | 0.280h-l |
| 31 | 1.04lm | 33.17d-i | 26ijkl | 2103.3l-o | 6781i-l | 0.363d-l |
| 32 | 1.14g-k | 32.75e-i | 36a-f | 1238qr | 6266j-m | 0.238l |
| 33 | 1.35ab | 40.52d | 33e-j | 2950.9g-j | 7610f-k | 0.389b-k |
| 34 | 1.28bcde | 35.85c-f | 30.5f-l | 1877n-q | 6892i-l | 0.309g-l |
| 35 | 1.2e-h | 33.15e-i | 29f-l | 2248.2k-o | 8671e-i | 0.29g-l |
| 36 | 1.08jkl | 38.7cde | 41.75a-d | 3276.5e-h | 9433c-f | 0.353e-l |
| Range | 0.96-1.35 | 22-70.47 | 23-43.75 | 929.5-5530.4 | 3581-13301 | 0.238-0.641 |
| Mean | 1.16 | 34.67 | 31.86 | 2948.69 | 7966.58 | 0.39 |
| LSD | 0.09 | 7.36 | 8.25 | 646.89 | 2374.8 | 0.147 |
Note: Mean values followed by similar letter(s) in each column for each trait at each location is not significantly different each other. LSD (5%): Least Significant Difference at P<0.05; DF: Days to Flowering; PFP: Pod Filling Period; DM: Days to Maturity; PH: Plant Height; NPB: Number of Primary Branch; NSB: Number of Secondary Branch; NPP: Number of Pod Per Plant; NSP: Number of Seed per Plant; NSPP: Number of Seed Per Pod; HSW: Hundred Seed Weight; SPE: Seed Production Efficiency; SY: Seed Yield; BY: Biomass Yield; HI: Harvest Index.
Table 4. Mean performance of 36 Kabuli chickpea genotypes.
Estimates of genetic parameters
Phenotypic and genotypic variance components and coefficients of variation: The value of phenotypic variance was higher than a genotypic variance for most traits studied, indicates the highest contribution of the environmental effects to the phenotypic variance in these traits and selection on phenotypic bases of these traits may not be effective for genetic improvement unless the environmental conditions are optimize. However, the difference is not high for number of seeds per pod and harvest index, indicates that the phenotypic expression of these traits was less influenced by environmental factors, and selection on phenotypic bases of these traits may be effective for genetic improvement. Dev et al., and Fasil reported similar results in Kabuli chickpea genetic variability studies [23,27].
The PCV value ranged from 5% to 71.7% for number of secondary branch and seed yield and GCV also ranged from 5.9 to 73 % for number of secondary branch and seed yield. High GCV and PCV values were observed for days to flowering, biomass yield, the number of seeds per plant, seed yield, number of primary branches, harvest index, hundred seed weight, and number of pod per plant, indicate the existence of wide genetic variation among the genotypes and had a possibility of genetic improvement through selection for these traits. The remaining studied traits except number of secondary branches exhibited moderate GCV values. The difference between GCV and PCV values was very low for most traits, indicating these traits are highly controlled by additive gene action and less influenced by the environment. In line with the present result, Joshi et al., reported high GCV and PCV values for biomass yield, seed yield, harvest index and 100 seed weight. Similarly, Awol et al., noted high GCV and PCV for seed yield, biomass yield, days to flowering, and days to maturity, number of pods per plant, number of seeds per pod and number of seeds per plant [2,41]. Mohan and Thiyagarajan [17] reported high GCV and PCV values for biomass yield, seed yield, and hundred seed weight. Zerfu et al., also reported high GCV and PCV for hundred seed weight, seed yield and biomass yield [46].
Broad sense heritability: The highest estimate of heritability values were observed for all studied traits, indicates selection based on phenotypic appearance of individual genotypes for such trait might be calm which infers these traits competency of replying to selection compression due to relatively small contribution of the environment to the phenotype. However, choosing superior individuals based on heritability evaluations alone may not be indication for genetic improvement. Similar findings reported by Dev et al., [23] Shengu et al., [47] and Fasil [27] for hundred seed weight, days to maturity, above ground biomass yield, harvest index, plant height, and number of primary branches, number of seed per plant and seed yield. Joshi et al., [2] reported high estimate of heritability for seed yield, biomass yield and hundred seed weight. However, the selection of superior individuals based on heritability estimates alone may not be an indication of genetic improvement. High heritability alone is not enough to make sufficient improvement through selection generally in advance generations unless accompanied by substantial amount of genetic advance [48]. The efficacy of heritability is increasing with the estimation of genetic advance, which indicates the degree of gain in a trait obtained under a particular selection pressure [49].
Genetic advance: Biomass yield (106.6), seed yield (147.7), number of primary branches (136.4), harvest index (80), hundred seed weight (72.6), number of seed per plant (34.5), number of pod per plant (93.3), number of seeds per plant (133), days to maturity (29.1), pod filling period (37.9), days to flowering (41.2) and plant height (29.8) had high GAM (Table 5). This indicates selection of genotype based on this trait might result in a high response in the new population. A genetic advance exceeding 100 percent for some studied traits indicates a substantial improvement in the trait and suggests that the selection process has been effective in promoting desirable genetic characteristics. Similarly, these results were agreed by Awol et al., [41] reported high GAM values for seed yield, biomass yield, number of pods per plant, and hundred seed weight. In the same way, Mohan and Thiyagarajan [17] reported high GAM for the number of secondary branches and number of pods per plant. In addition Zerfu et al., [46] reported high GAM values for the number of secondary branches, seed yield, biomass yield, hundred seed weight and harvest index. Higher heritability coupled with high GAM suggests that the traits are controlled by additive gene action. Days to flowering, biomass yield, and the number of seeds per plant, seed yield, and number of primary branches, harvest index, hundred seed weight, and number of pod per plant had high values of GCV, heritability and genetic advance as percentages of the mean. Such situations are the greatest potential initiated by additive genetic factors, there by imitating the effectiveness of selection for the enhancement of these traits. Therefore, these traits are significant for the genetic improvement of the Kabuli chickpea. Similar to this result high heritability coupled with high GAM values were reported for traits hundred seed weight, number of pods per plant, number of secondary branches, number of seeds per plant, seed yield and harvest index [27]. Thus, genetic advance is yet another important selection parameter that aids breeder in a selection programs. It has been emphasized that without genetic advance, the heritability values would not be of practical importance in selection based on phenotypic appearance. So, the genetic advance should be considered along with heritability in coherent selection breeding programs [50].
| Trait | Min-Max | Mean | σ 2e | σ2g | σ2gy | σ 2P | GCV | PCV | H2 | GA | GAM |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DF | 44.75-66.5 | 57.2 | 12.33 | 130.97 | 5.06 | 136.58 | 20 | 20.4 | 97.9 | 23.6 | 41.2 |
| PFP | 61.25-89 | 74.85 | 27.33 | 189.66 | 4.66 | 198.82 | 18.4 | 18.8 | 97.7 | 28.4 | 37.9 |
| DM | 110.25-148.75 | 130.11 | 35.56 | 337.34 | 6.26 | 349.36 | 14.1 | 14.4 | 98.3 | 37.8 | 29.1 |
| PH | 42.12-60.87 | 47.59 | 18.62 | 47.52 | 54.61 | 79.48 | 14.5 | 18.7 | 77.3 | 14.2 | 29.8 |
| NPB | 1.77-6 | 3.99 | 0.21 | 6.97 | 0.85 | 7.45 | 66.2 | 68.4 | 96.8 | 5.4 | 136.4 |
| NSB | 42.12-60.87 | 47.59 | 2 | 5.7 | 3.25 | 7.82 | 5 | 5.9 | 85.3 | 4.9 | 10.3 |
| NPP | 28.9-70.75 | 48.24 | 19.8 | 476.98 | 300.16 | 632.01 | 45.3 | 52.1 | 86.9 | 45 | 93.3 |
| NSP | 0.96-1.35 | 1.16 | 0.0041 | 0.04 | 0.01 | 0.05 | 17.8 | 19.1 | 93.4 | 0.4 | 34.5 |
| NSPP | 22-70.47 | 34.67 | 27.2 | 500.85 | 69.63 | 542.47 | 64.6 | 67.2 | 96.1 | 46.1 | 133 |
| HSW | 23-43.75 | 31.86 | 34.19 | 126.22 | 64.79 | 167.16 | 35.3 | 40.6 | 86.9 | 23.1 | 72.6 |
| SY | 929.5-5530.4 | 2948.69 | 209955.6 | 4469882 | 230656.2 | 4637699 | 71.7 | 73 | 98.2 | 4355.3 | 147.7 |
| BY | 3581-13301 | 7966.58 | 2829656 | 17008267 | 8999803 | 22215583 | 51.8 | 59.2 | 87.5 | 8495.7 | 106.6 |
| HI | 0.238-0.641 | 0.39 | 0.0108 | 0.02 | 0.03 | 0.04 | 39.1 | 51.8 | 75.5 | 0.3 | 80 |
Note:*: significant at (p ≤ 0.05) and **: highly significant (p ≤ 0.01) probability level.
CV: Coefficient of Variation, DF: Days to 50% Flowering; DM: Days to 90% Maturity;
PFP: Pod Filling Period; PH: Plant Height; NPB: Number of Primary Branches;
NSB: Number of Secondary Branches;
NPP: Number of Pods Per Plant; NSPP: Number of Seeds Per Plant;
NSP: Number of Seeds Per pod; BY: Biomass Yield;
HSW: Hundred Seed Weight; SY: Seed Yield and HI: Harvest Index.
Table 5. Estimates of genetic parameters for 13 traits of 36 Kabuli chickpea genotypes.
The degree of association between pairs of trait at the genotypic and phenotypic levels was estimated and results are presented in Table 5.
Genotypic correlation coefficients of seed yield with other traits: Designing, evaluating and setting selection criteria for the ideal characters in breeding program of crop plants are proceed from studies of correlation at the genotypic and phenotypic levels [51]. Genotypic correlation study provides the direction, strength and the point of association between the traits beyond the environmental effect. Seed yield showed positive and significant (P ≤ 0.05) genotypic association with the number of seeds per pod (r=0.41) and number of seed per plant (r=0.35). In agreement with this finding Tesfamichael, et al., [52] reported positive and significant (p<0.05) correlation of seed yield kg ha-1with biomass yield kgha-1(r=0.85), number of pods plant-1 (r=0.57), but non-significantly correlated with other traits such as days to 50% flowering, plant height, number of primary branches plant-1 [50]. Similarly, significant and positive association was reported among seed yield with number of pods plant-1, number of seed plant-1 by Fiaz S, et al. [53]. Tadesse, et al., [54] also reported significant and positive correlation of seed yield with biomass yield, plant height, number of pods per plant and number of seeds per pod, while other traits showed non-significant association with seed yield. Fasil, [27] reported highly significant and positive association of number of seeds per plant, hundred seed weight, and number of pods per plant with seed yield kgha-1.
Genotypic correlation coefficients of among other traits: Genotypic correlation coefficient among other related traits are shown in Table 5. Days to flowering had positive association with days to maturity (r=0.63), while highly and negatively significant genotypic correlation with number of seed per plant (r=-0.34). The result indicates genotypes that exhibited prolonged flowering period may result delaying of maturity, reducing number of seed per plant. Positive and highly significant association exhibited for days to maturity with pod filling period (r=0.76), this indicates early matured genotype took the shortest pod filling period. Similar to the present findings [17,27,55] reported positive and significant association of days to maturity with days to flowering and pod filling period. Number of primary branch showed positive and significant genotypic correlation with number of secondary branch (r=0.64), number of pod per plant (r=0.72) and negative significant association with biomass yield (r=-0.39). Number of Hasan and Deb [56] reported the same result for association of number of primary branch with number of pod per plant, number of seed per plant and number of secondary branch. Ali et al., [57] also reported positive and significant association of number of primary branches per plant with number of pods per plant and seeds per pod. Zerafu et al., [46] reported high and significant association between number of primary branch and secondary branch.
Number of seed per pod showed significant and positive genotypic association with number of number of seed per plant (r=0.599), 100 seed weight (r=0.51) and seed yield (r=0.41). Positive and significant association was recorded for number of seed per plant with seed yield (r=0.6), biomass yield(r=0.34) and harvest index (r= 0.41). Similarly, noted that highly significant and positive associations for number of seed per pod with number of pod per plant [42,51]. Above ground biomass yield showed significant and positive genotypic association with seed yield (r=0.51) and harvest index (r=0.48). Getachew et al., [42] and Awol and Asnake [58] revealed positive and significant association for above ground dry biomass yield with seed yield, while negative and significant association with harvest index. Tesfamichael et al., [52] also noted that biomass yield kgha-1 showed positive and significant (p<0.05) association with number of pods per plant-1 (r=0.75) and it was negatively and highly significant (p<0.01) association with 100 seed weight (r=-0.53).
Phenotypic correlation coefficients of seed yield with other traits: Correlation analysis indicated that seed yield exhibited significant and positive phenotypic associations with the pod filling period (r=0.16), number of pods per plant (r=0.17), number of seeds per pod (r=0.22), number of seeds per plant (r=0.16) and biomass yield (r= 0.52), indicates these traits are important for seed yield improvement. Yield is a polygenic traits controlled by several simply inherited traits. The correlation coefficients highlight the pattern of association among these yield components and helps in determining the important trait to improve the crop. In line with the present findings, similar results are reported in chickpea study [11,27,59].
Phenotypic correlation coefficients among other traits: Positive and significant association was recorded between days to flowering with days to maturity (r=0.54), while negative and highly significant association with number of seed per plant (r=0.32) and hundred seed weight (r=0.22). Positive and highly significant association exhibited for pod filing period with days to maturity (r=0.88), number of secondary branches (r=0.18), number of pods per plant (r=0.43). While, negative and significant association were observed pod filling period with number of seeds per pod and number of seed per plant, this indicates early pod filling genotypes were expected to be low number of seeds per pod and plant. Positive and significant correlation was recorded with number of pod per plant (r=0.48) and negative and significant association with number of seed per pod and plant. [37,56] and [27] reported similar association between traits.
Highly and positive phenotypic associations were recorded for number of seed per pod with number of seed per plant (r= 0.5), above ground biomass yield (r=0.34) and seed yield (r=0.22). Number of seed per plant showed significant and positive phenotypic association with seed yield (r=0.46), biomass yield (r=0.16), number of primary branches(r=0.17) and harvest index (r=0.25), while highly and negative significant association with days to flowering (r=-0.35), pod filling period (r=-0.28) and days to maturity (r=-0.39). Above ground biomass yield showed significant and positive phenotypic association with number of pod per plant (r=0.18), number of seed per pod (r=0.34), number of seed per plant (r=0.46) and seed yield (r=0.52) and harvest index (r=0.48). Similarly Tadesse et al. [54] and Amare [60] reported positive and significant association for above ground biomass yield with number of pod per plant, seed yield, number of seed per plant and number of seed per pod. Hundred seed weight showed negative and significant phenotypic correlation with days to flowering (r=-0.22), number of primary branches (r=-0.20). Positive and significant associations were recorded with plant height (r=0.186). Positive and significant association were observed for harvest index with number of seed per plant (r=0.25) and biomass yield(r=-0.48), but negative and significant association with seed yield (r=0.41). Plant height showed non-significant phenotypic association with all traits. Fasil [27] reported positive and significant correlation for harvest index with number of primary branch and number of seed per plant, contrary to the present result, plant height had significant phenotypic correlation with 100 seed weight [26].
Selection of genotypes for high yield alone is difficult because yield is the end product of components of several characteristics and has a polygenic inheritance. There might be component interactions in which a gene conditioning an increase in one trait will influence another provided other conditions are kept constant. Therefore, the selection of genotypes with high mean values for these traits along with high seed yield seems more important rather than selection for yield per se alone. Understanding the association of other traits with seed yield and the selection of genotypes for yield and for traits that have a significant correlation with yield is important. From the present result among 13 traits, the number of pods per plant, number of seeds per pod, number of seeds per plant and biomass yield had positive and significantly associated with seed yield at both genotypic and phenotypic levels. Therefore, these traits might be important selection criteria for further yield improvement of Kabuli chickpea.
Path coefficient analysis
In the present study seed yield was considered as a resultant variable and number of pods per plant, number of seeds per pod, number of seeds per plant and biomass yield, were causal (independent) variables in both genotypic and phenotypic (Tables 6 and 7).
| Variable | DF | PFP | DM | PH | NPB | NSB | NPP | NSP | NSPP | BY | HSW | GY | HI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DF | 1 | 0.02 | 0.63** | 0.15 | -0.002 | -0.075 | 0.035 | -0.145 | 0 | -0.24 | -0.18 | -0.24 | 0.031 |
| PFP | 0.081 | 1 | 0.76** | -0.055 | -0.069 | 0.14 | -0.062 | -0.21 | -0.13 | 0.28 | -0.042 | 0.129 | -0.27 |
| DM | 0.54** | 0.88** | 1 | 0.059 | -0.049 | 0.072 | -0.024 | -0.2524 | -0.34* | 0.065 | -0.138 | -0.04 | -0.19 |
| PH | 0.076 | 0.029 | 0.06 | 1 | 0.0628 | -0.22 | -0.053 | -0.0064 | -0.19 | 0.17 | 0.148 | 0.172 | -0.038 |
| NPB | 0.0108 | 0.04 | 0.0485 | 0.0451 | 1 | 0.68** | 0.72** | -0.0551 | 0.2898 | -0.387** | -0.041 | -0.08 | 0.0017 |
| NSB | -0.059 | 0.184* | 0.15 | -0.133 | 0.36** | 1 | 0.155 | -0.087 | -0.031 | -0.038 | -0.045 | -0.054 | 0 .053 |
| NPP | 0.174 | 0.433** | 0.48** | -0.085 | 0.47** | 0.155 | 1 | 0.042 | 0.26 | -0.316 | 0.13 | 0.074 | 0.072 |
| NSP | -0.15 | -0.20** | -0.24** | 0.015 | -0.035 | -0.087 | -0.1 | 1 | 0.599** | 0.062 | 0.51** | 0.41** | 0.21 |
| NSPP | -0.35** | -0.28** | -0.39** | -0.054 | 0.17* | -0.031 | -0.097 | 0.509** | 1 | -0.18 | 0.60** | 0.34* | 0.41** |
| HSW | -0.22** | 0.12 | -0.015 | 0.186* | -0.20** | -0.038 | -0.135 | 0.045 | -0.12 | 1 | -0.013 | 0.106 | -0.205 |
| BY | -0.1 | 0.05 | 0.013 | 0.1 | -0.055 | -0.044 | 0.18* | 0.34** | 0.46** | -0.027 | 1 | 0.51** | 0.48** |
| SY | -0.13 | 0.16* | 0.082 | 0.12 | -0.016 | -0.05 | 0.17* | 0.22** | 0.16* | 0.13 | 0.52** | 1 | -0.03 |
| HI | 0.012 | -0.14 | -0.1 | -0.011 | -0.057 | 0.053 | -0.037 | 0.095 | 0.25** | -0.16 | 0.48** | -0.41** | 1 |
Note: ***, **,* indicate very highly significant at 0.1%, highly significant at 1%, and significant at 5% probability levels, respectively. DF: Days to 50% Flowering; DM: Days to 90% Maturity; PFP: POD Filling Period; PH: Plant Height; NPB: Number of Primary Branches; NSB: Number of Secondary Branches; NPP: Number of Pods per Plant; NSP: Number of Seeds per Pod; NSPP: Number of Seeds Per Plant; BY: Biomass Yield; HI: Harvest Index; HSW: Hundred Seed Weight; SY: Seed Yield.
Table 6. Genotypic (above diagonal) and phenotypic (below diagonal) correlations coefficients of the 13 traits over years.
|
NSP | NSPP | BY | Rg |
|---|---|---|---|---|
| NSP | 0.44 | -0.57 | -0.25 | 0.27 |
| NSPP | -0.6 | 0.61 | -0.69 | -0.05 |
| BY | -0.14 | -0.65 | 0.79 | 0.64 |
Note: Residual = 0.46, NPP=Number of pods per plant, NSP=Number of seeds per pod, NSPP=number of seeds per plant and BY= biomass yield, rg=genotypic correlation coefficient
Table 7. Genotypic direct (bold and diagonal) and indirect effects of traits on seed yield for studied. Genotypes.
Genotypic direct and indirect effects of various characters on seed yield: Biomass yield (0.79), number of seeds per plant (0.61) and number of seed per pod (0.44) had a positive direct effect on seed yield. Therefore, these traits should be included in selection parameters for obtaining the maximum seed yield of Kabuli chickpea would be quite effective for the breeding program for yield improvement rather than for yield per se alone. The indirect effect of number of seeds per plant and number of seeds per pod on yield was negative via biomass yield. The indirect effect of biomass yield and number of seed per pod via number of seeds per plant exerted a negative effect on seed yield. In the same way, Tadesse et al. [54] reported the number of pods per plant increased seed yield indirectly through biomass yield. Agrawal et al., [55] and Fasil [27] also reported biomass yield and pods per plant had a high positive direct effect on seed yield. Shafique et al., [61] noted the positive direct effect of the number of pods per plant and the number of seeds per plant on seed yield. The residual effect of (0.46) indicates that the trait, which is included in the genotypic path analysis, explained 54 % of the total variations in seed yield.
Phenotypic direct and indirect effects of various traits on seed yield: Biomass yield showed a strong positive direct effect on seed yield (1.14). The number of seed per pod had positive direct effect on seed yield. While, negative direct effect had observed in number of seed per plant on seed yield. The indirect effect of number of seeds per pod had via number of seed per plant had been positive and negative for biomass yield. The direct effect of number of seeds per plant, biomass yield and number of seed per pod was positive. This indicates the importance of the traits to be used as direct selection criteria to improve seed yield. Similarly, Tadesse et al., [54] reported that the number of pods per plant increased seed yield indirectly through above ground biomass at the phenotypic level. Gizachew [11] reported days to maturity, number of pods per plant, number of seeds per plant, hundred seed weight and plant height had a positive direct effect on seed yield. The residual effect of (0.15) indicates that the trait, which is included in the phenotypic path analysis, explained 85 % of the total variation in seed yield. The remaining 15 % variation was the contribution of other factors, which is due to low environmental effects on the expression of the phenotypes (Table 8).
| NSP | NSPP | BY | Rg | |
|---|---|---|---|---|
| NSP | 0.39 | 0.03 | -0.16 | 0.27 |
| NSPP | -0.16 | -0.08 | 0.03 | -0.05 |
| BY | -0.04 | 0.03 | 0.14 | 0.64 |
Note: Residual=0.15, NPP=Number of Pods per Plant, NSP=Number of Seeds per Pod, NSPP=Number of Seeds Per Plant and BY=Biomass Yield and rp=phenotypic correlation coefficient
Table 8. Phenotypic direct (bold and diagonal) and indirect effects of traits on seed yield for studied genotypes
Conclusion
In conclusion, the analysis of variance showed the existence of high genetic diversity among the studied Kabuli chickpea genotypes. Combined analysis of variance showed significant variations for most traits. The role of gene action was high for the expression of most traits such as biomass yield, number of seeds per plant, seed yield, indicating the existence of wide genetic variation among the genotypes and had a possibility of genetic improvement through selection for these traits. High heritability coupled with high genetic advance as a percent of mean was observed for traits biomass yield, seed yield, number of seeds per pod, number of pods per plant showing that the high heritability is most likely due to additive gene effects; and the importance of selection for the improvement of Kabuli chickpea for these traits. Seed yield showed a highly significant and positive association at both genotypic and phenotypic levels with the number of seeds per pod, number of seeds per plant and biomass yield. Therefore, the association of these traits with seed yield is important for further Kabuli chickpea improvement. The number of seeds per pod, number of seeds per plant and biomass yield had a direct effect on seed yield increment. Hence, from the present results, it has been observed adequate existence of variability for most of the traits in the studied genotypes, which need to be, exploited in future Kabuli chickpea breeding. However, this study was conducted for two seasons and at one location which needs to be conducted in subsequent breeding trials considering more locations to develop high yielding varieties. It would be worthwhile to study qualitative traits among genotypes to identify accessions having high quality and marketable produce for progress improvement via the selection and/or hybridization program of Kabuli chickpea.
Data Availability
The original data used for data analysis and presented in the text. Further, inquiries can be directed to the corresponding author.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
The authors would like to express their great appreciation to the Ministry of Education, Debre Markos University, Debre Markos Branch Seed quality control Authority, Highland Pulse Research Program, DZARC and Awabel District Agricultural Office, for facilitating the research process.
Funding
This work was supported by the Ministry of Education Ethiopia, Debre Markos University, Debre Markos Branch Seed quality control Authority and self-supported.
About the Authors
Corresponding Author
Gebremeskel Mequanint Mulu
Department of Plant Science, College of Agriculture and Natural Resource, Debre Merkos University, P.O. Box-269, Debre Merkos, Ethiopia
References
- Merga B and Haji J (2019). Economic importance of chickpea: Production, value, and world trade. Cogent. Food. Agric. 5: 1615718.
- Joshi P, Yasin M and Sundaram P (2018). Genetic variability, heritability and genetic advance study for seed yield and yield component traits in a chickpea Recombinant Inbred Line (RIL) population. Int. J. Pure. Appl. Biosci. 6: 136-141.
- Muehlbauer FJ and Sarker A (2017). Economic importance of chickpea: production, value, and world trade. The. Chickpea. Genome. 5-12.
- Van der Maesen LJ (1987). Origin, history and taxonomy of chickpea. In The chickpea. 11-34.
- Wani SP, Chander G, Patil MD, Sawargavkar G and Kumar S (2021). Scaling Up Food Legume Production Through Genetic Gain and Improved Management. In: Saxena, K.B., Saxena, R.K., Varshney, R.K. (eds) Genetic Enhancement in Major Food Legumes. Springer, Cham.
- Fikre A and Bekele D (2019). Chickpea breeding and crop improvement in Ethiopia: Past, present and the future. Univers. J. Agric. Res. 8: 33-40.
- Rawal V and Navarro DK (2019). The global economy of pulses.
- Wallace TC, Murray R and Zelman KM (2016). The nutritional value and health benefits of chickpeas and hummus. Nutrients. 8: 766.
[Crossref] [Google Scholar] [PubMed]
- International Trade Centre (2019). Ethiopia Pulses strategy.
- CSA (2021). Area and production for major crops. Private Peasant Holdings, Meher Season 2020/2021 (2013 E.C.). Volume I, Statistical bulletin 590, Addis Ababa, Ethiopia.
- The Asbestos Disease Awareness Organization (ADAO) (2018). 2018 ADAO Asbestos Awareness and Prevention Conference Agenda. May 3rd, 2018.
- Atnaf M, Tesfaye K and Dagne K (2015). The importance of legumes in the Ethiopian farming system and overall economy: An overview. Am. J. Exp. Agric. 7: 347-358.
- Rashid S, Yirga C, Behute B and Lemma S (2010). Pulses value chain in Ethiopia: Constraints and opportunities for enhancing exports. International Food Policy Research Institute. Washington, DC, USA. 1.
- Getachew T. (2019). Pulse crops production opportunities, challenges and its value chain in Ethiopia: A review article. J Environ. Earth. Sci. 9.
- Millan T, Clarke HJ, Siddique KH, Buhariwalla HK, Gaur PM, et al. (2006). Chickpea molecular breeding: New tools and concepts. Euphytica. 147: 81-103.
- Kulsum U, Sarker U and Rasul MG (2022). Genetic variability, heritability and interrelationship in salt-tolerant lines of T. Aman rice. Genetika. 54: 761-776.
- Mohan S and Thiyagarajan K (2019). Genetic variability, correlation and path coefficient analysis in chickpea (Cicer arietinum L.) for yield and its component traits. Int. J. Curr. Microbiol. Appl. Sci. 8: 1801-1808.
- Hasan JM, Kulsum UM, Majumder RR and Sarker U (2020). Genotypic variability for grain quality attributes in restorer lines of hybrid rice. Genetika. 52: 973-989.
- Sarker U (2020). Variability, heritability, character association, and path coefficient analysis in advanced breeding lines of rice (Oryza sativa L.). Genetika. 52: 711-726.
- Hill WG and Mackay TF (2004). DS Falconer and Introduction to quantitative genetics. Genetics. 167: 1529-1536.
[Crossref] [Google Scholar] [PubMed]
- Dewey DR and Lu K (1959). A correlation and path‐coefficient analysis of components of crested wheatgrass seed production. Agronomy J. 51: 515-518.
- Thormann CE, Ferreira ME, Camargo LE, Tivang JG, et al. (1994). Comparison of RFLP and RAPD markers to estimating genetic relationships within and among cruciferous species. Theor. Appl. Genet. 88: 973-980.
[Crossref] [Google Scholar] [PubMed]
- Dev A, Verma P and Kumhar BL (2017) Genetic character variability studies in desi chickpea (Cicer arietinum L.) genotypes. Int. J. Curr. Microbiol. Appl. Sci. 6: 20-25.
- Singh SP (2001). Broadening the genetic base of common bean cultivars: A review. Crop. Sci. 41: 1659-1675.
- Chakrabarty T, Sarker U, Hasan M and Rahman MM (2018). Variability in mineral compositions, yield and yield contributing traits of stem amaranth (Amaranthus lividus). Genetika. 50: 995-1010.
- Ali Y, Atta BM, Akhter J, Monneveux P, et al. (2008). Genetic variability, association and diversity studies in wheat (Triticum aestivum L.) Germplasm. Pak. J. Bot. 40: 2087-2097.
- Hailu F (2020). Genetic variability, heritability and genetic advance of Kabuli chickpea (Cicer arietinum L.) for agronomic traits at Central Ethiopia. Int. J. Plant. Breed. Crop Sci. 7: 710-714.
- Yilma Kebede G (2000). Genetic variability and associations of yield and yield related traits in Kabuli chickpea (Cicer arietinum L.) Genotypes at Arsi-Robe, Southeastern Ethiopia Doctoral dissertation, Haramaya University.
- Prakash V (2006). Divergence analysis in kabuli chickpea (Cicer arietinum L.). Indian J. Genet. Plant. Breed. 66: 241-242.
- Ferede S, Yirga C, Kehaliew A, Agegnehu G, et al. (2020). Farming systems characterization and analysis in East Gojjam zone: Implications for Research and Development (R and D) interventions. Ethiop. Inst. Agric. Res.
- IBPGR I (1993). ICARDA, Descriptor for chickpea (Cicer arietinum L.).
- Upadhyaya HD, Gowda CL and Sastry DV (2008). Plant genetic resources management: Collection, characterization, conservation and utilization. J. SAT. Agric. Res. 6: 16.
- Gomez KA (1984). Statistical procedures for agricultural research. John NewYork: Wiley and Sons. 1984.
- Burton GW and de Vane DE (1953). Estimating heritability in tall fescue (Festuca arundinacea) from replicated clonal material.
- Deshmukh SN, Basu MS and Reddy PS (1986). Genetic variability, character association and path coefficients of quantitative traits in Virginia bunch varieties of groundnut.
- Allard RW (1999). Principles of Plant Breeding. John Wiley and Sons.
- Johnson PL and NANDA RS (2015) Genetic diversity and association analysis for yield traits chickpea (Cicer arietinum L.) under rice based cropping system. The Bioscan. 10: 879-884.
- Miller PA, Williams JC Jr, Robinson HF and Comstock RE (1985). Estimates of genotypic and environmental variances and covariances in upland cotton and their implications in selection 1. Agronomy. J. 50: 126-31.
- Wright S (1921). Correlation and causation. J. Agri. Res. 20: 557.
- Singh RK and Chaudhary BD (1981). Biometrical methods in quantitative genetic analysis.
- Mohammed A, Tesso B, Ojiewo C and Ahmed S (2019). assessment of genetic variability and heritability of agronomic traits of Ethiopian chickpea (Cicerarietinum L) Landraces. Black. Sea. J. Agri. 2: 10-15.
- Tilahun G, Mekbib F, Fikre A and Eshete M (2015) Genetic divergence and character association of Kabuli type chickpea (Cicer arietium) genotype under rainfed conditions in Ethiopia. Curr. Res. Agri. Sci. 2: 123-131.
- Vijayakumar AG, Boodi I, Gaur PM and Upadhyaya HD (2017). Genetic diversity for yield and its component traits in chickpea (Cicer arietinum L.). Electron. J. Plant Breed. 8: 89-95.
- Kebede GY and Haile GA. Genetic diversity analysis of Kabuli chickpea (Cicer arietinum L.) genotypes at Arsi-Robe, Southeastern Ethiopia.
- Malik SR, Ghulam Shabbir GS, Muhammad Zubir MZ, Iqbal SM, et al. (2014). Genetic diversity analysis of morpho-genetic traits in desi chickpea (Cicer arietinum).
- Zerfu A, Hailu F and Adal M (2021). Phenotypic variability and association (among yield components) and yield related trait in Desi type chickpea (Cicer arietinum L.) in Raya Kobo district, Northern Ethiopia. J. Agric. Sci. Agrotechnol. 10.
- Sarker U, Islam MT, Rabbani MG and Oba S (2014). Genotypic variability for nutrient, antioxidant, yield and yield contributing traits in vegetable amaranth. J. Food. Agric. Environ. 12: 168-174.
- Keles d, Ozgen S, Saracoglu O, Ata A, Ozgen M (2016). Antioxidant potential of Turkish pepper (Capsicum annuum L.) genotypes at two different maturity stages. Turk. J. Agric. For. 40: 542-551.
- Shengu MK, Hirpa D and Wolde Z (2018). Genetic variability of some chickpea (Cicer arietinum L.) genotypes and correlation among yield and related traits in humid tropics of southern Ethiopia. J. Plant. Breed. Crop. Sci.
- Sarker U, Islam T, Rabbani G and Oba S (2015) Genotype variability in composition of antioxidant vitamins and minerals in vegetable amaranth. Genetika. 47: 85-96.
- Johnson HW, Robinson HF, Comstock RE (1995). Estimates of genetic and environmental variability in soybeans.
- Tesfamichael SM, Githiri SM, Nyende AB and Rao NV (2015). Variation for agro-morphological traits among Kabuli chickpea (Cicer arietinum L.) genotypes. J. Agri. Sci. 7: 75-92.
- Fiaz S, Aslam M, Wattoo FM, Riaz A, Bashir I (2015). Interrelationships among yield and yield contributing traits in chickpea (Cicer arietinum L.). Bangladesh. J. Plant. Breed. Genet. 28: 39-46.
- Tadesse M, Fikre A, Eshete M, Girma N, Korbu L, et al. (2016). Correlation and path coefficient analysis for various quantitative traits in desi chickpea genotypes under rainfed conditions in Ethiopia. J. Agri. Sci. 8: 112-118.
- Agrawal T, Kumar A, Kumar S, Kumar A, Kumar RR, et al. (2018). Correlation and path coefficient analysis for grain yield and yield components in chickpea (Cicer arietinum L.) under normal and late sown conditions of Bihar, India. Int. J. Curr. Microbiol. App. Sci. 7: 1633-1642.
- Hasan MT and Deb AC (2017). Assessment of genetic variability, heritability, character association and selection indexes in chickpea (Cicer arietinum L.). Int. J. Biosci. 10: 111-129.
- Ali Q, Tahir MH, Sadaqat HA, Arshad S, Farooq J, et al. (2011) Genetic variability and correlation analysis for quantitative traits in chickpea genotypes (Cicer arietinum L.). J. Bacteriol. Res. 3: 6-9.
- Amare T (2019). Genetic variability and association of traits for Desi type chickpea (Cicer arietinum L.) advanced lines under potential environment in North Gondar, Doctoral dissertation, Ethiopia
- Kayan N and Adak MS (2012). Associations of some characters with grain yield in chickpea (Cicer arietinum L.). Pak. J. Bot. 44: 267-272.
- Shafique MS, Muhammad Ahsan MA, Zafar Mehmood ZM, Muhammad Abdullah MA, et al. (2016). Genetic variability and interrelationship of various agronomic traits using correlation and path analysis in chickpea (Cicer arietinum L.).
- Adem Am and Fikre A (2018). Correlation and path coefficient analysis among seed yield and yield related traits of Ethiopian chickpea (Cicer arietinum L.) landraces. Acta Agric Slovenica. 111: 661-670.
Keywords:
Download:
Full PDF- Share This