Hierarchical outcrossing among and within fruits in Bertholletia excelsa Bonpl. (Lecythidaceae) open-pollinated seeds
Received: December 24, 2017
Accepted: January 18, 2018
Published: January 31, 2018
Genet.Mol.Res. 17(1): gmr16039872
DOI: 10.4238/gmr16039872
Abstract
Knowing the mating patterns is important to determine the number of trees necessary for seed collection for conservation ex situ, tree breeding and environmental reforestation purposes. We investigated B. excelsa individuals and fruits, to check mating system index variations in a population by using open-pollinated seeds which were hierarchically sampled within and among fruits from nine trees genotyped for ten microsatellite loci. Outcrossing rate ( )changed between trees (0.49–1.0) and fruits (0.53–1.0), but seeds were predominantly produced by outcrossing (0.92) at mean population level. Mating between related trees ( ) was detected in six trees (0.04–0.08) and in 32 fruits in trees (0.03–0.22), thus it suggests that the population presented some related trees in our intra-population spatial genetic structure. Individual fixation index values of seed trees ( ) were lower than the seedling fixation index values ( ), fact that suggests the selection against inbred individuals between the seed and adult stages. The correlated mating showed that seeds sampled at population level were predominantly composed of half-sibs (66%) and full-sibs (20%). Paternity correlation was significantly higher within ( ) than among ( ) fruits at population level, mostly in individual trees. Results evidenced that mating was not random due to self-fertilization, to mating between related trees and correlated mating, and families comprised different relatedness levels such as half-sibs, full-sibs, self-sibs and self-half-sibs; some cases, mating presented inbreeding. These results were addressed to discuss strategies for seed collection applied to conservation ex situ, tree breeding and environmental reforestation.
Introduction
Non-timber forest products (NTFPs) from native forests contribute to biodiversity conservation (Arnold and Ruiz-Pérez, 1998; Marshall et al., 2005) in the Amazon region, since their exploitation generates income and encourages forest preservation by rural communities in order to improve the quality of life of local producers (Bayama et al., 2014). Thus, it is necessary to reconcile the productivity and ecological sustainability of this tree species to generate NTFP incomes (Wadt et al., 2008).
Bertholletia excelsa Bonpl. (Lecythidaceae), popularly known as Brazil nut, presents seeds with high commercial and nutritional value (Wadt et al., 2015; Baldoni et al., 2017). Brazil nut (NTFP) extraction favors forest preservation; therefore, it is considered a model species for conservation if one also takes into account that wood cutting is prohibited by law (Wadt et al., 2005). The species is distributed in the Amazon region, which is known by its diversity of plant species presenting the potential to be used in agriculture and in tree breeding. However, the lack of knowledge about the species, associated with deforestation, agricultural frontier advancements and wildfire events, contributes to natural population losses, even before they are studied (Maués and Oliveira, 2010).
Understanding species mating systems concerns knowing its composition and the genetic structure of its populations (Richards, 1997; Luna et al., 2005). The mating system is the way individuals, populations or species recombine their genetic variability in each generation in order to generate offspring. Being aware of such system is relevant for population genetic conservation management, for tree breeding and environmental reforestation plans (Sebbenn, 2006). Overall, species can be classified by mating system: i) autogamous, when outcrossing is ≤ 0.2; ii) outcross, when it is ≥ 0.8; iii) mixed mating systems, when outcrossing is between 0.2 and 0.8 (Goodwille et al., 2005). B. excelsa is classified as an outcrossing species and its floral structures preclude self-fertilization, as well as limit the group of animals capable of reaching pollen in their flowers (O'Malley et al., 1988; Maués, 2002; Cavalcante, 2008). Their main pollinating agents are bees belonging to genus Bombus, Xylocopa and Centris (Müller, 1995; Maués, 2002). Flowers house the reproductive organs in a chamber (ula) and these groups of bees are able to reach the pollen, since their physical vigor and robust bodies make the task feasible (Maués, 2002). Despite being an outcrossing species, some studies observed selffertilization in Brazil nut at population and individual level (O'Malley et al., 1988; Wadt et al., 2015; Baldoni et al., 2017).
Molecular markers such as microsatellite loci or SSR (Simple Sequence Repeats) have the advantage of being codominant, heritable and very polymorphic. Moreover, they amplify large numbers of alleles (Hoshino et al., 2002) and represent low cost in investigations about the genetic diversity and mating system of tree species when the primers were already developed, such as in B. excelsa. Reis et al. (2009) developed 12 polymorphic microsatellite markers to the species and Sujii et al. (2013) developed another 12. In addition, genetic studies with molecular markers in fragmented forests using natural seeds or regenerates helped evidencing processes such as genetic drift, gene flow, selection and mating system (Carvalho et al., 2010; Wadt et al., 2015; Baldoni et al., 2017).
Although it is forbidden to cut B. excelsa trees, the strong deforestation in the Amazon region and the illegal logging already extinguished many of its populations; therefore, conservation strategies are required. Knowing the mating system of a tree species is important for genetic conservation, tree breeding and environmental reforestation, since fertilization patterns determine the relatedness and inbreeding of next generations and, consequently, the effective size of seed collection (Sebbenn 2006). The mating system of B. excelsa has been investigated (O'Malley et al. 1988, Wadt et al. 2015), as well as its pollen dispersal patterns (Baldoni et al. 2017). These studies have been investigating the outcrossing rates, the mating between related trees, the paternity correlation among and within fruit variations, and pollen-dispersal distance and patterns. We investigated population and individual outcrossing rate variations, and paternity correlation among and within fruits in order to add information about fruit within tree mating system. We tested the following hypotheses: i) Are seeds in population, individual seed trees and fruit levels produced by outcrossing? As the floral structures of the species preclude self-fertilization, we expected all seeds to be originated from outcrossing; ii) is there mating between relatives at population and individual level, as well as within fruits? There are reports about populations presenting intra-population spatial genetic structure; therefore, we expected to record mating between relatives at all these levels; iii) Is the paternity correlation within fruits higher than among fruits? We expected to find this pattern because of reports in previous studies about higher paternity correlation within than among fruits in the species.
Materials and Methods
Study site and sampling
The study was conducted in a 9ha permanent plot (717971.64 E, 8774887.57 S) located in legal reserve at Santo Ângelo Farm, which belongs to the Dalpai Group. The farm is located approximately 30 km from Itaúba County, Mato Grosso State, Brazil, where seed extraction is performed in a yearly basis. The plot is located in a natural Amazon forest. The Bertholletia excelsa population density was 19.8 trees/ha and presented diameter at breast height > 10 cm. Nine trees were selected in the center of the experimental plot; fruits were hierarchically collected from them. Seeds were germinated in sandbox under 50% shade after their tegument was removed at Embrapa Agrossilvipastoril, Sinop, MT. This procedure generated 300 progenies duly identified based on their origin (seed tree and fruit). The plant material from all adult trees (trunk cambium) and progenies (leaves) was collected for DNA extraction. The leaves were transported in plastic bags containing silica gel and stored in a freezer (-20° C). The trunk cambium was stored in 1 ml of transport buffer (300 μl of 2% CTAB buffer, 700 μl of absolute ethanol, 0.2 g of ascorbic acid) and stored in cold chamber (4° C).
Microsatellite analysis
The vascular cambium and leaf of the trunk were used to extract the total DNA from the samples based on the methodology described by Doyle and Doyle (1990), with modifications (CTAB from 2 to 4%). Polymerase Chain Reaction (PCR) amplification was carried out in ten microsatellite loci: BET12, BET14, BET15, BET16, BEX02, BEX09, BEX22, BEX27, BEX33, and BEX37, as described by Reis et al. (2009), Sujii et al. (2013) and Cabral et al. (2017). These analyses were conducted at the Genetics Laboratory of Embrapa Genetic Resources and Biotechnology, Brasília, DF. Fragment sizes were identified in base pairs in the GeneMapper 4.1® software (Applied Biosystems). Data extracted from the GeneMapper software were rounded out in the Allelobin software (Idury and Cardon, 1997).
Mating system analysis
The mating system at population, individual seed tree and fruits within tree level was estimated through mixed and correlated mating models that were assessed in the MLTR 3.1 software based on the Expectation Maximization Numerical method (Ritland, 2002). The estimated indices were pollen and ovules gene frequencies, seed trees fixation index 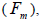 multilocus
multilocus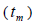 and single-locus
and single-locus 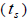 outcrossing rates, mating between relatives
outcrossing rates, mating between relatives 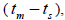 selfing correlation
selfing correlation 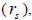 multilocus paternity correlation within and among fruits (rp); within
multilocus paternity correlation within and among fruits (rp); within  and among
and among 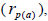 fruits. Fruits presenting only one seed were excluded from the analyses conducted to estimate
fruits. Fruits presenting only one seed were excluded from the analyses conducted to estimate  and
and 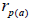 at population and individual seed tree level, only fruits with at least three seeds were used in the analyses. Standard deviation indices at 95% confidence intervals were calculated by using 1,000 bootstraps as re-sampling units between individuals within a single family. The effective number of pollen donors was estimated among and within fruits
at population and individual seed tree level, only fruits with at least three seeds were used in the analyses. Standard deviation indices at 95% confidence intervals were calculated by using 1,000 bootstraps as re-sampling units between individuals within a single family. The effective number of pollen donors was estimated among and within fruits 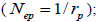 within fruits
within fruits 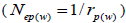 and, among fruits
and, among fruits 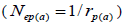 based on Ritland (1989). The proportion of pairwise self-sibs
based on Ritland (1989). The proportion of pairwise self-sibs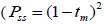 , half-sibs
, half-sibs 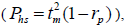 full-sibs
full-sibs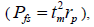 self-half-sibs
self-half-sibs and mean coancestry coefficient within progeny
and mean coancestry coefficient within progeny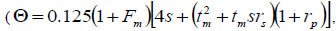 wherein s is the selfing rate:
wherein s is the selfing rate: 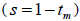 followed the methodology by Sebbenn (2006). Seed trees individual fixation index (Fm) was estimated in the SPAGEDI software (Hardy and Vekemans, 2002). The variance effective size (Ne) and fixation index (Fo) within a single family were estimated according to Wadt et al. (2015). Negative Fm and Fo values were assumed to be zero in and
followed the methodology by Sebbenn (2006). Seed trees individual fixation index (Fm) was estimated in the SPAGEDI software (Hardy and Vekemans, 2002). The variance effective size (Ne) and fixation index (Fo) within a single family were estimated according to Wadt et al. (2015). Negative Fm and Fo values were assumed to be zero in and  and Ne estimates (Wadt et al., 2015). The number of trees required for collecting seeds for conservation purposes was estimated through
and Ne estimates (Wadt et al., 2015). The number of trees required for collecting seeds for conservation purposes was estimated through 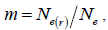 wherein:
wherein: 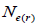 is the required effective population size (Sebbenn, 2006), which was herein assumed to be 150 (Lacerda et al., 2008). The 95% CI applied to the indices was described according to Wadt et al. (2015). The spearman correlation coefficient (ρ) was used to investigate whether there was significant association between sample size (n) pairwise variables within families, and within fruits,
is the required effective population size (Sebbenn, 2006), which was herein assumed to be 150 (Lacerda et al., 2008). The 95% CI applied to the indices was described according to Wadt et al. (2015). The spearman correlation coefficient (ρ) was used to investigate whether there was significant association between sample size (n) pairwise variables within families, and within fruits,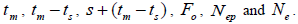
Results
The mean population fixation index of seed trees 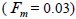 was low, but significantly higher than zero, thus it
indicated an inbreeding process; however, it was negative at individual level and ranged from -0.71 to -0.37
(Table 1). The mean population fixation index of seedlings was similar to zero
was low, but significantly higher than zero, thus it
indicated an inbreeding process; however, it was negative at individual level and ranged from -0.71 to -0.37
(Table 1). The mean population fixation index of seedlings was similar to zero 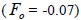 but ranged from -
0.42 to 0.12 between families (Table 1) and from -0.43 to 0.34 (Table 2) between tree fruits. All Fo values were
higher than Fm (Table 1), and it suggested the selection against inbred individuals between the seed and adult stages. The mean outcrossing rate
but ranged from -
0.42 to 0.12 between families (Table 1) and from -0.43 to 0.34 (Table 2) between tree fruits. All Fo values were
higher than Fm (Table 1), and it suggested the selection against inbred individuals between the seed and adult stages. The mean outcrossing rate 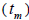 at population level (0.92) was not significantly different from the unity (1.0), but, at individual level, it was significantly lower than 1.0 in five seed trees (it ranged from 0.49 to 0.98) (Table 1) and in fruits within trees in three fruits (it ranged from 0.53 to 0.79) (Table 2). Self-fertilization correlation
at population level (0.92) was not significantly different from the unity (1.0), but, at individual level, it was significantly lower than 1.0 in five seed trees (it ranged from 0.49 to 0.98) (Table 1) and in fruits within trees in three fruits (it ranged from 0.53 to 0.79) (Table 2). Self-fertilization correlation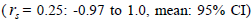 was not significantly different from zero. Mating between
related trees
was not significantly different from zero. Mating between
related trees 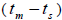 was significantly higher than zero at individual level in six trees (it ranged from 0.04 to
0.08) and it ranged from 0.03 to 0.22 in 32 fruits within trees (Tables 1 and 2).
was significantly higher than zero at individual level in six trees (it ranged from 0.04 to
0.08) and it ranged from 0.03 to 0.22 in 32 fruits within trees (Tables 1 and 2).
| Tree | n |  |
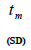 |
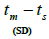 |
 |
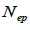 |
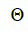 |
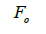 |
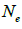 |
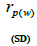 |
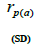 |
 |
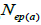 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 78 | -0.62 | 0.88 (0.05) | 0.08 (0.03) | 0.04 (0.02) | 22.7 | 0.162 | 0.07 | 2.99 | 0.28 (0.14) | 0.03 (0.02) | 3.5 | 34.5 |
| 2 | 10 | -0.61 | 0.98 (0.00) | 0.06 (0.01) | 0.23 (0.39) | 4.3 | 0.157 | -0.42 | 2.61 | 0.07 (0.62) | 0.29 (0.47) | 13.5 | 3.4 |
| 3 | 31 | -0.71 | 0.97 (0.02) | 0.05 (0.02) | 0.11 (0.14) | 9.3 | 0.144 | -0.14 | 3.21 | 0.40 (0.34) | 0.06 (0.16) | 2.5 | 16.9 |
| 4 | 37 | -0.46 | 1.00 (0.01) | 0.04 (0.01) | 0.53 (0.12) | 1.9 | 0.191 | 0.05 | 2.50 | 0.94 (0.11) | 0.46 (0.13) | 1.1 | 2.2 |
| 5 | 22 | -0.57 | 0.49 (0.12) | -0.34 (0.10) | 0.62 (0.27) | 1.6 | 0.304 | 0.08 | 1.59 | 1.00 (0.26) | 0.48 (0.38) | 1.0 | 2.1 |
| 6 | 31 | -0.50 | 0.63 (0.11) | -0.18 (0.08) | 0.35 (0.17) | 2.9 | 0.251 | 0.12 | 1.91 | 0.41 (0.36) | 0.34 (0.19) | 2.5 | 2.9 |
| 7 | 35 | -0.48 | 1.00 (0.00) | 0.05 (0.01) | 0.02 (0.04) | 52.6 | 0.127 | 0.02 | 3.62 | 0.37 (0.21) | 0.21 (0.05) | 2.7 | 4.8 |
| 8 | 41 | -0.42 | 1.00 (0.00) | 0.05 (0.01) | 0.13 (0.05) | 7.5 | 0.142 | -0.16 | 3.32 | 0.53 (0.17) | 0.08 (0.06) | 1.9 | 12.3 |
| 9 | 15 | -0.37 | 0.97 (0.03) | 0.03 (0.03) | 0.09 (0.20) | 11.0 | 0.144 | -0.08 | 2.99 | 0.36 (0.57) | 0.01 (0.35) | 2.8 | 142.9 |
| Mean | 33.3 | 0.03 | 0.92 | 0.00 | 0.23 | 4.3 | 0.178 | -0.07 | 2.34 | 0.67 | 0.18 | 1.5 | 5.6 |
| 95% CI | - | 0.02/0.04 | 0.86/1.00 | 0.00/0.01 | 0.04/0.41 | 2.4/23.8 | 0.133/0.230 | -0.18/0.04 | 1.90/2.88 | 0.37/0.83 | 0.01/0.30 | 1.2/2.7 | 3.4/66.7 |
n is the sample size; Fm and Fo are the fixation indices of seed trees and seedlings, respectively; tm is the multilocus
outcrossing rate; tm - ts is the rate of mating between relatives; 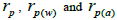 are the multilocus correlation of paternity among, and within, and within and among fruits, respectively;
are the multilocus correlation of paternity among, and within, and within and among fruits, respectively;  are the effective numbers of pollen donors among, and within; and within and among fruits, respectively;
are the effective numbers of pollen donors among, and within; and within and among fruits, respectively; 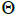 is the coancestry coefficient within progeny; Ne is the effective size variance; SD is the standard deviation; 95% confidence intervals are in parentheses (95% CI).
is the coancestry coefficient within progeny; Ne is the effective size variance; SD is the standard deviation; 95% confidence intervals are in parentheses (95% CI).
Table 1. Inbreeding and mating system indices at individual and mean population level for Bertholletia excelsa
| Tree | Fruit | n | 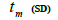 |
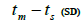 |
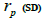 |
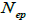 |
 |
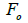 |
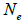 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 6 | 0.96 (0.09) | 0.06 (0.08) | 0.47 (0.20) | 2.1 | 0.189 | -0.03 | 2.07 |
| 3 | 4 | 0.99 (0.01) | 0.08 (0.01) | 0.10 (0.08) | 10.3 | 0.139 | -0.07 | 2.18 | |
| 5 | 7 | 0.97 (0.07) | 0.09 (0.07) | 0.12 (0.15) | 8.3 | 0.146 | -0.09 | 2.55 | |
| 6 | 6 | 0.96 (0.08) | 0.05 (0.07) | 0.10 (0.01) | 10.2 | 0.146 | 0.04 | 2.40 | |
| 8 | 3 | 0.99 (0.01) | 0.05 (0.01) | 0.28 (0.17) | 3.6 | 0.162 | -0.29 | 1.82 | |
| 9 | 10 | 1.00 (0.00) | 0.07 (0.01) | 0.10 (0.03) | 10.2 | 0.138 | -0.11 | 2.87 | |
| 10 | 6 | 0.85 (0.15) | -0.01 (0.11) | 0.11 (0.12) | 9.0 | 0.176 | -0.09 | 2.17 | |
| 11 | 4 | 0.97 (0.06) | 0.03 (0.05) | 0.10 (0.01) | 10.2 | 0.145 | -0.21 | 2.14 | |
| 12 | 7 | 1.00 (0.00) | 0.05 (0.01) | 0.07 (0.08) | 13.3 | 0.135 | -0.22 | 2.68 | |
| 13 | 6 | 0.93 (0.13) | 0.01 (0.11) | 0.09 (0.01) | 11.2 | 0.153 | -0.19 | 2.37 | |
| 14 | 6 | 0.94 (0.11) | 0.04 (0.09) | 0.08 (0.02) | 12.8 | 0.148 | -0.12 | 2.42 | |
| 16 | 5 | 0.97 (0.07) | 0.09 (0.07) | 0.09 (0.01) | 10.8 | 0.144 | -0.09 | 2.32 | |
| 2 | 3 | 3 | 1.00 (0.00) | 0.06 (0.04) | 0.05 (0.05) | 20.8 | 0.131 | -0.35 | 1.97 |
| 4 | 4 | 1.00 (0.00) | 0.00 (0.00) | 0.05 (0.05) | 20.8 | 0.131 | -0.43 | 2.24 | |
| 3 | 1 | 6 | 1.00 (0.00) | 0.05 (0.01) | 0.12 (0.22) | 8.3 | 0.140 | -0.29 | 2.50 |
| 2 | 3 | 1.00 (0.00) | 0.04 (0.01) | 0.08 (0.02) | 12.0 | 0.136 | -0.40 | 1.94 | |
| 3 | 3 | 1.00 (0.00) | 0.03 (0.01) | 0.26 (0.27) | 3.9 | 0.158 | -0.09 | 1.84 | |
| 4 | 3 | 1.00 (0.00) | 0.07 (0.02) | 0.07 (0.02) | 15.2 | 0.133 | -0.02 | 1.96 | |
| 7 | 4 | 0.86 (0.35) | -0.03 (0.23) | 0.18 (0.23) | 5.5 | 0.180 | -0.05 | 1.92 | |
| 8 | 4 | 1.00 (0.00) | 0.03 (0.01) | 0.46 (0.30) | 2.2 | 0.183 | -0.29 | 1.91 | |
| 9 | 5 | 0.79 (0.18) | -0.17 (0.00) | 0.08 (0.02) | 13.0 | 0.189 | -0.09 | 1.99 | |
| 4 | 1 | 4 | 0.53 (0.28) | 0.22 (0.17) | 0.08 (0.02) | 12.7 | 0.273 | 0.34 | 1.34 |
| 3 | 3 | 1.00 (0.00) | 0.05 (0.01) | 0.50 (0.19) | 2.0 | 0.187 | -0.06 | 1.71 | |
| 6 | 7 | 1.00 (0.00) | 0.03 (0.01) | 0.89 (0.15) | 1.1 | 0.237 | 0.18 | 1.82 | |
| 7 | 7 | 1.00 (0.00) | 0.01 (0.01) | 0.98 (0.13) | 1.0 | 0.248 | -0.37 | 1.74 | |
| 8 | 4 | 1.00 (0.00) | 0.06 (0.02) | 0.09 (0.02) | 11.5 | 0.136 | -0.13 | 2.20 | |
| 9 | 6 | 1.00 (0.00) | 0.05 (0.03) | 0.08 (0.02) | 12.5 | 0.135 | 0.32 | 2.25 | |
| 5 | 2 | 3 | 0.95 (0.35) | 0.05 (0.28) | 0.76 (0.10) | 1.3 | 0.224 | 0.09 | 1.51 |
| 6 | 6 | 1.00 (0.00) | 0.01 (0.01) | 0.10 (0.16) | 10.3 | 0.137 | -0.10 | 2.53 | |
| 7 | 6 | 1.00 (0.00) | 0.06 (0.02) | 0.00 (0.07) | 1.0 | 0.125 | 0.00 | 2.67 | |
| 6 | 1 | 5 | 1.00 (0.10) | 0.04 (0.07) | 0.04 (0.09) | 28.6 | 0.130 | -0.12 | 2.45 |
| 6 | 6 | 1.00 (0.00) | 0.01 (0.01) | 0.15 (0.18) | 6.9 | 0.143 | -0.01 | 2.47 | |
| 9 | 4 | 1.00 (0.11) | 0.09 (0.08) | 0.94 (0.04) | 1.1 | 0.243 | 0.16 | 1.53 | |
| 10 | 4 | 1.00 (0.00) | 0.05 (0.02) | 0.48 (0.31) | 2.1 | 0.185 | -0.28 | 1.90 | |
| 7 | 1 | 4 | 1.00 (0.00) | 0.04 (0.01) | 0.16 (0.21) | 6.1 | 0.146 | 0.03 | 2.10 |
| 2 | 7 | 1.00 (0.03) | 0.06 (0.03) | 0.10 (0.04) | 9.7 | 0.138 | 0.10 | 2.54 | |
| 3 | 3 | 1.00 (0.00) | 0.03 (0.01) | 0.40 (0.24) | 2.5 | 0.175 | -0.29 | 1.77 | |
| 5 | 4 | 1.00 (0.00) | 0.06 (0.01) | 1.00 (0.39) | 1.0 | 0.250 | -0.03 | 1.60 | |
| 6 | 5 | 1.00 (0.00) | 0.05 (0.03) | 0.08 (0.04) | 12.8 | 0.135 | 0.02 | 2.38 | |
| 7 | 4 | 1.00 (0.00) | 0.08 (0.03) | 0.20 (0.22) | 5.1 | 0.150 | -0.27 | 2.11 | |
| 8 | 4 | 1.00 (0.00) | 0.03 (0.01) | 0.25 (0.26) | 4.1 | 0.156 | 0.08 | 1.99 | |
| 8 | 3 | 4 | 1.00 (0.00) | 0.03 (0.01) | 0.37 (0.26) | 2.7 | 0.172 | -0.27 | 1.97 |
| 4 | 9 | 1.00 (0.00) | 0.01 (0.01) | 0.50 (0.28) | 2.0 | 0.187 | -0.43 | 2.25 | |
| 5 | 3 | 1.00 (0.00) | 0.07 (0.02) | 0.27 (0.21) | 3.7 | 0.159 | -0.29 | 1.83 | |
| 7 | 6 | 1.00 (0.00) | 0.04 (0.02) | 0.09 (0.04) | 10.8 | 0.137 | -0.20 | 2.54 | |
| 8 | 3 | 0.70 (0.26) | -0.21 (0.00) | 0.09 (0.03) | 10.1 | 0.217 | -0.09 | 1.61 | |
| 9 | 4 | 1.00 (0.00) | 0.06 (0.03) | 0.07 (0.02) | 14.5 | 0.134 | -0.07 | 2.22 | |
| 10 | 7 | 1.00 (0.00) | 0.12 (0.05) | 0.22 (0.19) | 4.5 | 0.153 | -0.09 | 2.47 | |
| 9 | 4 | 6 | 0.83 (0.21) | 0.04 (0.17) | 0.24 (0.31) | 4.1 | 0.191 | -0.07 | 2.06 |
n is the sample size; tm is the multilocus outcrossing rate; tm - ts is the mating rate between relatives; rp, is the mating rate between relatives; correlation of paternity within fruits; Nep is the effective number of pollen donors within fruits; 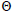 is the coancestry coefficient; Fo is the fixation index of seedlings; Ne is the variance effective size; SD is the standard deviation.
is the coancestry coefficient; Fo is the fixation index of seedlings; Ne is the variance effective size; SD is the standard deviation.
Table 2. Inbreeding and mating system indices at individual fruit level for fruits with at least three seeds
Paternity correlation 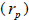 indicates that 23.0% of outcrossing seedlings at population level were full-sibs; it
ranged from 0.02 to 0.62 between seed trees, and from 0 to 1.0 between fruits within trees. Thus, 4.3 pollen
donors effectively
indicates that 23.0% of outcrossing seedlings at population level were full-sibs; it
ranged from 0.02 to 0.62 between seed trees, and from 0 to 1.0 between fruits within trees. Thus, 4.3 pollen
donors effectively  fertilized seed trees, on average; fertilization ranged from 1.6 to 52.6 between trees
and from 1.0 to 28.6 within fruits (Tables 1 and 2). On average, 0% (95% CI: 0-2%) of the seedlings were selfsibs
fertilized seed trees, on average; fertilization ranged from 1.6 to 52.6 between trees
and from 1.0 to 28.6 within fruits (Tables 1 and 2). On average, 0% (95% CI: 0-2%) of the seedlings were selfsibs 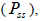 66% (95% CI: 44-97%) were half-sibs
66% (95% CI: 44-97%) were half-sibs 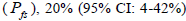 were full-sibs
were full-sibs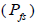 and 14% (95% CI: 0-27%) were self-half-sibs
and 14% (95% CI: 0-27%) were self-half-sibs at population level. Accordingly, the coefficient of coancestry
at population level. Accordingly, the coefficient of coancestry 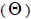 at population and individual level, and within fruits, was higher; the variance effective size
at population and individual level, and within fruits, was higher; the variance effective size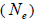 within a single
family was lower than the expected in panmictic populations
within a single
family was lower than the expected in panmictic populations 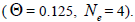 The number of seed trees for
seed collection (m) was estimated in 64 (95% CI: 52-79). Paternity correlation was significantly higher within
fruits
The number of seed trees for
seed collection (m) was estimated in 64 (95% CI: 52-79). Paternity correlation was significantly higher within
fruits 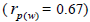 than among fruits
than among fruits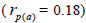 at population level, and higher within fruits
at population level, and higher within fruits than
among fruits
than
among fruits 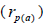 in eight of the nine seed trees.
in eight of the nine seed trees.
The sample size  was not significantly correlated to the mating system indices at family level, but it was
significantly associated with
was not significantly correlated to the mating system indices at family level, but it was
significantly associated with  at fruit within trees level (Table 3). Therefore, we excluded
the fruits with less than three seeds in order to decrease sample size variation between fruits within trees.
Subsequently, only n and Ne was significantly associated
at fruit within trees level (Table 3). Therefore, we excluded
the fruits with less than three seeds in order to decrease sample size variation between fruits within trees.
Subsequently, only n and Ne was significantly associated 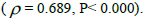 The indices
The indices 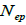 and Ne had significant positive association at family and fruit level, and the index
and Ne had significant positive association at family and fruit level, and the index 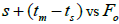 had significant
positive association at fruit level.
had significant
positive association at fruit level.
| Family level | Fruit level | |||
|---|---|---|---|---|
 |
P | 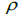 |
P | |
 |
0.295 | 0.441 | 0,220 | 0.063 |
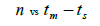 |
0.346 | 0.361 | -0.261 | 0.030 |
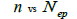 |
0.276 | 0.472 | 0.029 | 0.807 |
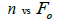 |
0.218 | 0.574 | -0.249 | 0.034 |
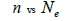 |
0.370 | 0.327 | 0.853 | 0.000 |
 |
0.281 | 0.464 | 0.127 | 0.383 |
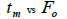 |
-0.621 | 0.074 | -0.202 | 0.163 |
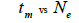 |
0.628 | 0.070 | 0.143 | 0.328 |
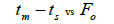 |
-0.553 | 0.125 | 0.257 | 0.074 |
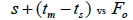 |
0.462 | 0.210 | 0.332 | 0.020 |
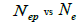 |
0.828 | 0.006 | 0.521 | 0.000 |
Table 3. Results of Spearman correlation coefficient ( ) and statistical probability (P) between sample size (n) pairwise variables, tm , tm - ts , Nep [
) and statistical probability (P) between sample size (n) pairwise variables, tm , tm - ts , Nep [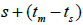 ], Fo and Ne
], Fo and Ne
Discussion
Outcrossing rate
Our results indicate individual and fruit outcrossing rate  variation, and tm lower than the unity (1.0) in five seed trees (0.49 - 0.98), as well as in three fruits within trees (0.53 - 0.79). Therefore, B. excelsa overall produces seeds through outcrossing; however, there are some variations between tree and fruits within trees, due to individual variations in self-incompatibility or due inbreeding depression; thus, same trees may produce seeds from self-fertilization. The species presents floral structure capable of precluding self-fertilization (O'Malley et al., 1988; Maués, 2002; Cavalcante, 2008), although self-fertilization at population level (at range 0.85 to 0.98) and at individual level (at range 0.90 to 0.98) have been reported in other studies (O'Malley et al., 1988; Wadt et al., 2015; Baldoni et al., 2017). These results suggest that the species presents latent self-incompatibility; some self-fertilization events may result in seed production. An alternative explanation for tm variation between trees and fruits lies on genetic load population and on individual variations that result in the survival of self seeds originated from populations, or from trees, with low genetic load (Hufford and Hamrick, 2003; Tambarussi et al., 2016). Our results also evidenced that outcrossing can vary between fruits within trees; thus, some openpollinated seeds are inbred due to self-fertilization.
variation, and tm lower than the unity (1.0) in five seed trees (0.49 - 0.98), as well as in three fruits within trees (0.53 - 0.79). Therefore, B. excelsa overall produces seeds through outcrossing; however, there are some variations between tree and fruits within trees, due to individual variations in self-incompatibility or due inbreeding depression; thus, same trees may produce seeds from self-fertilization. The species presents floral structure capable of precluding self-fertilization (O'Malley et al., 1988; Maués, 2002; Cavalcante, 2008), although self-fertilization at population level (at range 0.85 to 0.98) and at individual level (at range 0.90 to 0.98) have been reported in other studies (O'Malley et al., 1988; Wadt et al., 2015; Baldoni et al., 2017). These results suggest that the species presents latent self-incompatibility; some self-fertilization events may result in seed production. An alternative explanation for tm variation between trees and fruits lies on genetic load population and on individual variations that result in the survival of self seeds originated from populations, or from trees, with low genetic load (Hufford and Hamrick, 2003; Tambarussi et al., 2016). Our results also evidenced that outcrossing can vary between fruits within trees; thus, some openpollinated seeds are inbred due to self-fertilization.
Mating among related trees
Our results also showed mating between related individuals 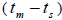 in six trees (from 0.04 to 0.08) and within fruits (from 0.03 to 0.22). According to these results, the population presents some related trees, due to the possible occurrence of intra-population spatial genetic structures (SGS). Related individuals were often closer than the unrelated ones, due to short seed dispersal and closeness to the mother. SGS was detected in two populations living 175 m from each other, thus it indicated that near-neighbor trees living at this distance from each other may be related (Baldoni et al., 2017). Other study also detected mating between relatives (from 0.02 to 0.08), and it is explained by the fact that the mean pollen dispersal distance (159 m) lies within the distance SGS occurs in (Wadt et al., 2015); it may also explain our
in six trees (from 0.04 to 0.08) and within fruits (from 0.03 to 0.22). According to these results, the population presents some related trees, due to the possible occurrence of intra-population spatial genetic structures (SGS). Related individuals were often closer than the unrelated ones, due to short seed dispersal and closeness to the mother. SGS was detected in two populations living 175 m from each other, thus it indicated that near-neighbor trees living at this distance from each other may be related (Baldoni et al., 2017). Other study also detected mating between relatives (from 0.02 to 0.08), and it is explained by the fact that the mean pollen dispersal distance (159 m) lies within the distance SGS occurs in (Wadt et al., 2015); it may also explain our 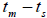 results. Furthermore, it also reinforces that the species presented self-incompatibility variation between trees in our genetic load and that inbreeding may also due to mating between related trees.
results. Furthermore, it also reinforces that the species presented self-incompatibility variation between trees in our genetic load and that inbreeding may also due to mating between related trees.
Inbreeding
Our results showed that  has positive association at fruit level. Thus, the increased selfing or mating between related trees increased inbreeding within fruits. The individual fixation index values of seed trees
has positive association at fruit level. Thus, the increased selfing or mating between related trees increased inbreeding within fruits. The individual fixation index values of seed trees  were lower than the seedling fixation index values
were lower than the seedling fixation index values and it suggested the selection against inbred individuals between the seed and adult stages. Thus, inbreed seeds originated from selfing and mating between related trees will probably die before reaching the adult stage due to inbreeding depression. Inbreeding between the seeds in adult stage is apparently a common pattern in tropical trees, as it has been reported in many studies (Hufford and Hamrick, 2003; Degen and Sebbenn, 2014; Wadt et al., 2015; Tambarussi et al., 2016).
and it suggested the selection against inbred individuals between the seed and adult stages. Thus, inbreed seeds originated from selfing and mating between related trees will probably die before reaching the adult stage due to inbreeding depression. Inbreeding between the seeds in adult stage is apparently a common pattern in tropical trees, as it has been reported in many studies (Hufford and Hamrick, 2003; Degen and Sebbenn, 2014; Wadt et al., 2015; Tambarussi et al., 2016).
Correlated mating
The sampled seeds were predominantly composed of half-sibs (66%) and full-sibs (20%) at population level. The mean population paternity correlation was 0.23, but it varied between seed trees (0.02-0.62), and between fruits within trees (0 to 1.0). Thus, in mean population level, a low number of pollen donors effectively (Nep = 4.3) fertilized the seed trees; it ranged from 1.6 to 52.6 between trees, and from 1.0 to 28.6 within fruits. The maximum estimated Nep of 28.6 is an obvious overestimate, because a single fruit produces from 8 to 24 seeds. Our Nep results at population level are similar to those recorded for other populations (4.5) distributed in natural forests (Wadt et al., 2015). The paternity correlation within fruits 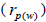 was higher than between fruits
was higher than between fruits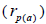 at population level
at population level 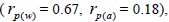 as well as at individual level (Table 1). Thus, the number of effective pollen donors was often lower within
as well as at individual level (Table 1). Thus, the number of effective pollen donors was often lower within  than between fruits
than between fruits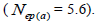 Wadt et al. (2015) detected similar pattern in other populations of this species
Wadt et al. (2015) detected similar pattern in other populations of this species  higher than
higher than 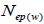 is also reported in many studies involving insect-pollinated tree species (Muona et al., 1991; Sampson, 1998; Quesada et al., 2001; Tamaki et al., 2009; Silva et al., 2011; Manoel et al., 2015; Tambarussi et al., 2015).
is also reported in many studies involving insect-pollinated tree species (Muona et al., 1991; Sampson, 1998; Quesada et al., 2001; Tamaki et al., 2009; Silva et al., 2011; Manoel et al., 2015; Tambarussi et al., 2015).
CONCLUSION AND IMPLICATIONS FOR SEED COLLECTION
Our results evidenced that mating was not random due to self-fertilization, through mating between related trees
and correlated mating. Consequently, families comprised different relatedness levels such as self-sibs, half-sibs,
full-sibs, and self-half-sibs; eight of them presented inbreeding from selfing, from 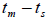 or from both. Such
results are the key to determine the strategies to collect seeds for conservation ex situ, tree genetic breeding and environmental reforestation purposes. The coancestry coefficient
or from both. Such
results are the key to determine the strategies to collect seeds for conservation ex situ, tree genetic breeding and environmental reforestation purposes. The coancestry coefficient at population and individual level, as
well as within fruits, was higher due to the mix of relatedness and inbreeding within families; the variance effective size (Ne) within a single family was lower than the expected for panmictic populations
at population and individual level, as
well as within fruits, was higher due to the mix of relatedness and inbreeding within families; the variance effective size (Ne) within a single family was lower than the expected for panmictic populations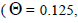
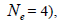 fact that resulted in at least 64 seed trees for seed collection. However, as there were variations in the
outcrossing rate, in mating between related trees and in correlated mating within fruits. We suggest that seed
collection must involve many fruits from each seed tree. Seeds from different seed trees must be mixed, first.
Subsequently, they should be mixed with seeds from other trees at the same proportion to maternal gametic
controls; therefore, all mothers will contribute with the same number of genes.
fact that resulted in at least 64 seed trees for seed collection. However, as there were variations in the
outcrossing rate, in mating between related trees and in correlated mating within fruits. We suggest that seed
collection must involve many fruits from each seed tree. Seeds from different seed trees must be mixed, first.
Subsequently, they should be mixed with seeds from other trees at the same proportion to maternal gametic
controls; therefore, all mothers will contribute with the same number of genes.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgment
The authors are grateful for the research support granted by Embrapa (Empresa Brasileira de Pesquisa Agropecuária), CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), and FAPEMAT (Fundação de Amparo à Pesquisa do Estado de Mato Grosso). Alexandre M. Sebbenn and Flávio D. Tardin were granted with the research fellowship from CNPq.
About the Authors
Corresponding Author
A.B. Baldoni
Empresa Brasileira de Pesquisa Agropecuária (Embrapa), Embrapa Agrossilvipastoril, Rodovia dos Pioneiros MT-222, Km 2,5, CEP 78550-970, Sinop, Mato Grosso, Brazil
- Email:
- diazcastro@unemat.br
References
- Arnold JEM and Ruiz-Pérez M (1998). The role of nontimber forest products in conservation and development. In: Wollenberg E, Ingles A. (Eds.): Incomes from the forest: methods for the development and conservation of forest products for local communities: 17-42. CIFOR/IUCN, Bogor.
- Baldoni AB, Wadt LHO, Campos T (2017). Contemporary pollen and seed dispersal in natural populations of Bertholletia excelsa (Bonpl.). Genetics and Molecular Research 16(3):gmr16039756. https://doi.org/10.4238/gmr16039756
- Bayama MMA, Malavazi FW, Sá CP (2014). Aspectos da cadeia produtiva da castanha-do-Brasil no estado do Acre, Brasil. Boletim do Museu Paraense Emílio Goeldi. Ciências Naturais 9(2):417-426.
- Brondani RPV and Grattapaglia D (2001). Cost-effective method to synthesize a fluorescent internal DNA standard for automated fragment sizing. Biotechniques 31:802-810. https://doi.org/10.1007/bf02752700
- Cabral JC, Baldoni AB, Tonini H (2017). Diversity and genetic structure of the native Brazil nut tree Bertholletia excelsa Bonpl.) population. Genetics and Molecular Research 16(3):gmr16039702. https://doi.org/10.4238/gmr16039702
- Carvalho ACM, Freitas MLM, Moraes SMB (2010). Diversidade genética, endogamia e fluxo gênico em pequeno fragmento florestal de óleo de copaíba (Copaifera langsdorffii Desf.). Revista Brasileira de Botânica 44:599-606.
- Cavalcante MC (2008). Visitantes florais e polinização da castanha-do-brasil (Bertholletia excelsa H.&B.) em cultivo na Amazônia central. Master’s thesis, UFC, Fortaleza.
- Degen B and Sebbenn AM (2014). Genetic and tropical forest. In: Tropical Forestry Handbook (Pancel L and Kölh M, eds.). 2nd edn. Springer Verlag, Berlin Heidelberg, 1-30.
- Doyle JJ and Doyle JL (1990). Isolation of plant DNA from fresh tissue. Focus 12:13-15.
- Finkeldey R (1998). An introduction to tropical forest genetics. Lecture Notes. Göttingen: Institute of Forest Genetics and Forest Tree Breeding, pp. 225.
- Freeland JR (2005). Molecular ecology. Sussex, Wiley.
- Fryxell P (1957). Mode of reproduction of higher plants. Botanical Review 23:135-233. https://doi.org/10.1007/bf02869758
- Hardy O and Vekemans X (2002). SPAGeDI: a versatile computer program to analyze spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618-620. https://doi.org/10.1046/j.1471-8286.2002.00305.x
- Hoshino AA, Palmieri DA, Bravo JP (2002). Marcador microssatélites na conservação de germoplasma vegetal. Biotecnologia, Ciência e Desenvolvimento 29:146-150.
- Hufford KM and Hamrick JL (2003). Viability selection at three early life stages of the tropical tree, Platypodium elegans (Fabaceae, Papilionoideae). Evolution 57:518-526. https://doi.org/10.1554/0014-3820(2003)057[0518:vsatel]2.0.co;2
- Idury RM and Cardon LR (1997). A simple method for automated allele binning in microssatellite markers. Genome Research 7:1104-1109. https://doi.org/10.1101/gr.7.11.1104
- Karasawa MMG, Dornelas MC, Araújo ACG, Oliveira GCX (2009). Biologia e genética dos sistemas reprodutivos. In: Karasawa MMG (Ed.). Diversidade Reprodutiva de Plantas. Ribeirão Preto: SBG, 26-52.
- Lacerda EBL, Sebbenn AM, Kanashiro M (2008). Long-pollen movement and deviation of random mating in a low-density continuous population of Hymenaea courbaril in the Brazilian Amazon. Biotropica 40:462–470. https://doi.org/10.1111/j.1744-7429.2008.00402.x
- Luna R, Epperson BK, Oyama K (2005). Spatial genetic structure of two sympatric neotropical palms with contrasting life histories. Heredity 95:298-305. https://doi.org/10.1038/sj.hdy.6800655
- Manoel RO, Freitas MLM, Furlani Junior E (2015). Individual, fruit, and annual variation in correlated mating in a Genipa americana population. Silvae Genetica 64:108–116. https://doi.org/10.1515/sg-2015-0010
- Maués MM (2002). Reproductive phenology and pollination of the Brazil nut tree (Bertholletia excelsa Humb. & Bompl. Lecythidaceae) in Eastern Amazonia. Pp. 245-254. In: P. Kevan, V.L. Imperatriz-Fonseca (eds.). Pollinating Bees – The conservation Link Between Agriculture and Nature. Ministry of Environment, Brasília, DF, 313p.
- Maués MM and Oliveira PEAM (2010). Conseqüências da fragmentação do habitat na ecologia reprodutiva de espécies arbóreas em florestas tropicais, com ênfase na Amazônia. Oecologia Australis 14(1):238-250.
- Marshall E, Newton AC, Schreckenberg K (2005). Commercialization of non-timber forest products: first steps in analysing the factors influencing success. International Forestry Review 5(2):128-137. https://doi.org/10.1505/ifor.5.2.128.17410
- Müller CH, Figueiredo FJC, Kato AK (1995). A castanha-do-Brasil. Brasilia: EMBRAPA/SPI, 65p.
- Muoana O, Moran GF, Bell JC (1991). Hierarchical patterns of correlated mating in Acacia melanoxylon. Genetics 127:619-626.
- O’Malley DM, Buckley DP, Prance GT (1988). Genetics of Brazil nut (Bertholletia excelsa Humb. & Bonpl.: Lecythidaceae). Theoretical and Applied Genetics 76:929-932.
- Quesada M, Fuchs E, Lobo J (2001). Pollen load size, reprocuctive success and progeny kinship of natural pollinated flowers of the tropical dry forest tree, Pachira quinta. Am J Bot 88:2113-2118. https://doi.org/10.2307/3558436
- Reis AMM, Braga AC, Lemes MR (2009). Development and characterization of microssatélites markers for the Brazil nut tree (Bertholletia excelsa) Humb. & Bonpl. (Lecytidaceae). Molecular Ecology Resources 9(3):920-923.
- Richards AJ (1997). Plant breeding systems. 2º ed. Cambridge: Chapmann e Hall, 529p.
- Ritland K (1989). Correlated matings in the partial selfer Mimulus guttatus. Evolution 43:848-859. https://doi.org/10.2307/2409312
- Ritland K (2002). Extensions of models for the estimation of mating systems using n independent loci. Heredity 88:221-228. https://doi.org/10.1038/sj.hdy.6800029
- Sampson JF (1998). Multiple paternity in Eucalyptus rameliana (Myrtaceae). Heredity 81:349-355.
- Sebbenn AM (2006). Sistema de reprodução em espécies arbóreas tropicais e suas implicações para a seleção de árvores matrizes para reflorestamentos ambientais. In: Higa AR, Silva LD. Pomares de sementes de espécies florestais nativas. Curitiba: FUPEF, p. 93-138.
- Silva CRS, Albuquerque PSB, Ervedosa FR (2011). Understanding the genetic diversity, spatial genetic structure and mating system at the hierarchical levels of fruits and individuals of a continuous Theobroma cacao population from the Brazilian Amazon. Heredity 106:973– 985. https://doi.org/10.1038/hdy.2010.145
- Silva VS, Martins K, Campos T (2012). Diversidade genética de populações naturais de castanheira (Bertholletia excelsa) com marcadores ISSR. In: Congresso Brasileiro de Recursos Genéticos, 2012, Belém, PA. Anais. Brasília, DF: Sociedade Brasileira de Recursos Genéticos, 2012. http://ainfo.cnptia.embrapa.br/digital/bitstream/item/71859/1/24499.pdf.
- Sujii PS, Inglis PW, Ciampi AY (2013). Isolation and characterization of microsatellite markers for Bertholletia excelsa (Lecythidaceae) population genetic analysis. Genetics and Molecular Research 12(4):5278-5282. https://doi.org/10.4238/2013.november.7.2
- Sujii PS, Martins K, Wadt LHO (2015). Genetic structure of Bertholletia excelsa populations from the Amazon at different spatial scales. Conservation Genetics 16:955-964. https://doi.org/10.1007/s10592-015-0714-4
- Tamaki I, Setsuko S, Tomaru N (2009). Estimation of outcrossing rates at hierarchical levels of fruit, individuals, populations and species in Magnolia stellate. Heredity 102:381-388. https://doi.org/10.1038/hdy.2008.128
- Tambarussi EV, Boshier D, Vencovsky R (2015). Several small: how inbreeding affects conservation of Cariniana legalis Mart. Kuntze (Lecythidaceae) the Brazilian Atlantic Forest's largest tree. International Forestry Review 18:502-510. https://doi.org/10.1505/146554816820127550
- Tambarussi EV, Boshier D, Vencovsky R (2016). Inbreeding depression from selfing and mating between relatives in the Neotropical tree Cariniana legalis Mart. Kuntze. Conservation Genetics 17:1-10. https://doi.org/10.1007/s10592-016-0896-4
- Wadt LHO, Kainer KA, Gomes-Silva DAP (2005). Population structure and nut yield of a Bertholletia excelsa stand in Southwestern Amazonia. Forest Ecology and Management 211:371-384. https://doi.org/10.1016/j.foreco.2005.02.061
- Wadt LHO, Kainer KA, Staudhammer CL (2008). Sustainable forest use in Brazilian extractive reserves: Natural regeneration of Brazil nut in exploited populations. Biological Conservation 141:322-346. https://doi.org/10.1016/j.biocon.2007.10.007
- Wadt LHO, Baldoni AB, Silva VS (2015). Mating system variation among populations, individuals and within and among fruits in Bertholletia excelsa. Silvae Genetica 64:5-6. https://doi.org/10.1515/sg-2015-0023
- Zanettini MHB and Cavalli SS (2003). Variabilidade Genética em Função do Modo de Reprodução. In: Freitas LB, Bered F (Eds.) Genética e Evolução Vegetal. Porto Alegre: Editora da UFRGS, 177-188.
Keywords:
Download:
Full PDF- Share This