High concentrations of sacharose lead to metabolic changes that favor the formation of co-products in ethanolic fermentation by Zymomonas mobilis
Received: December 24, 2017
Accepted: September 01, 2018
Published: November 05, 2018
Genet.Mol.Res. 17(4): http://dx.doi.org/gmr16039929
DOI: http://dx.doi.org/10.4238/gmr16039929
Abstract
Z. mobilis has attracted great biotechnology interest in recent decades due to its unique metabolism and the ability to produce bioethanol from simple sugars in an effectively and quickly way. Z. mobilis also produce other bioactive products as fructose, gluconic acid, sorbitol, levan, fructo-oligosaccharides. This study aimed to identify proteins differentially expressed by Z. mobilis ZAG-12 under fermentative conditions with low and high concentration of sucrose. Z. mobilis ZAG-12 was grown on SSDL in two fermentation conditions involving low (50g/L; 5%) or high (200g/L; 20%) sucrose concentrations. The bacteria were grown at 30°C without agitation under oxygen-limited conditions and cell growth was monitored by OD for 72 h. Sucrose, glucose, fructose, ethanol, sorbitol and levan were quantified in the fermentative liquid. The identification of differently expressed proteins were analyzed by Two-dimensional gel electrophoresis. The results showed that Z. mobilis ZAG-12 altered its protein expression and formation of products, such as ethanol and sorbitol, in response to a change in sucrose concentration. This response was characterized by proteomic analysis that detected 39 differentially expressed proteins, most related to transport and carbohydrate metabolism, amino acid metabolism and translation. The proteomic profile provided a better understanding of the physiological responses on how this promising microorganism redirects its metabolic flux to the production of ethanol and co-products.
Keywords
Sorbitol; Levan; Growth kinetics; Metabolic pathways.
Introduction
Faced with the crisis in the petrochemical sector, industry leaders begun to increase its search for alternative resources, aiming for the production of renewable energy sources such as bioethanol (Mohd Azhar and Abdulla, 2017). This scenario has driven several measures to increase ethanol production, such as the use of new substrates, selection of super productive microbial strains, as well as the optimization of cultivation conditions (Rostagno and Prado, 2015; Koti and Govumoni, 2016; Pena-Castro and Del Moral, 2017). The gram-negative bacteria Z. mobilis has attracted great biotechnology interest in recent decades due to its unique metabolism and the ability to produce bioethanol from simple sugars in an effectively and quickly way (Wang and Cao, 2016; Yang and Mohagheghi, 2016; Liu and Hsieh, 2017). Other important products produced by Z. mobilis include fructose (Zhang and Chen, 2009), gluconic acid (Erzinger and Vitolo, 2006), sorbitol (Erzinger and Vitolo, 2006; Liu and Dong, 2010), levan (Silbir and Dagbagli, 2014), fructo-oligosaccharides (Vigants and Upite, 2013) and various enzymes with biotechnological relevance (Aziz and Michlmayr, 2011; Lee and Hong, 2014; Goderska and Juzwa, 2015; Einsfeldt and Baptista, 2016). Z. mobilis also presents antagonistic activity toward pathogenic microorganisms and it has ability to modulate the immune response of the host as shown in a murine model of sepsis (Campos and Ximenes, 2013).
Given this biotechnological potential, the Department of Antibiotics from the Federal University of Pernambuco (UFPEDA) maintains a collection of Z. mobilis strains, most of them isolated as a fermentation contaminant (Silva and Araujo, 2015). Several genetic studies have been carried out with these strains including genome sequencing, recombinant strains production (De Morais and Rios, 1993; Kang and Kang, 1998; S. Lima, 2011; Kouvelis and Teshima, 2014; Silva and Araujo, 2015) and phylogenetic studies (Silva and Araujo, 2015). In one of these works, the Z. mobilis ZAG-12 strain (UFPEDA 241) was obtained by fusion the spheroplasts from two strains: (i) ZAP (UFPEDA 205), a levan producer strain; and (ii) Ag 11 (UFPEDA98) which do not produce levan (Morais 1984/85). This experiment was carried out in order to produce clones with high temperature tolerance (>38C), high capacity to produce ethanol (such as ZAP) and, consequently, low levan production, as it has been reported that levan biosynthesis can directly impact ethanol yield (Ananthalakshmy and Gunasekaran 1999; Lobato 2003). ZAG-12 (and other strains) has genetic characteristics different from the parental lineages (Silva, Araujo et al. 2015). In spite of these biotechnological aptitudes, little is known of the mechanisms involved in the metabolic regulation of this lineage in stress situations.
During fermentation, Z. mobilis is subjected to several physiological, ethanol and osmotic stresses. Proteomic strategies have been applied to better understand the mechanisms involved in the Z. mobilis response to ethanol stress (Yang, Pan et al. 2013) and its tolerance to sodium acetate (Yang and Franden, 2014). The adaptation to these factors has been shown to be complex and multifaceted, involving proteins of different functional categories. Chaperones, enzymes from glycolytic and fermentative pathway proteins and proteins involved in ATP synthesis are some examples (Yang and Pan, 2013; Yang and Pan, 2014; Mohagheghi and Linger, 2015).
In contrast, the mechanisms involved in Z. mobilis tolerance of high sucrose concentrations are not yet totally understood. It is known that when cultivated in high sucrose concentrations, Z. mobilis directs its metabolism to produce sorbitol, oligosaccharides and levan (Tano and Buzato, 2000; Sootsuwan and Thanonkeo, 2013). This condition may favor osmotic stress. However, the mechanisms that regulate the adaptation of this bacterium to this microenvironment are not yet known.
In the present study, we sought to identify proteins differentially expressed under fermentative conditions with low and high concentration of sucrose, using two-dimensional electrophoresis technology followed by MALDI-TOF mass spectrometry analysis, with the aim of providing knowledge about the cellular adaptation strategy developed by the unique Z. mobilis ZAG-12 strain.
Material and Methods
Bacterial lineage and growth conditions
Z. mobilis ZAG-12 (UFPEDA 241), belonging to the Microbial Collection of the Department of Antibiotics of the Federal University of Pernambuco (UFPEDA), was used. The strain was stored in SSDL medium containing 15% glycerol at -80°C. For the pre-inoculum, Z. mobilis ZAG-12 was grown on SSDL (Standard de Swings & De Ley) medium and incubated for 24 h. Two fermentation conditions involving low (50 g/L; 5%) or high (200 g/L; 20%) sucrose concentrations were established. The other components of the medium were not altered: yeast extract (5 g/L), KH2PO4 (2.0 g/L); MgSO4.7H2O (0.5 g/L); (NH4)2SO4 (1.0 g/L) and pH (6.5). The pre-inoculum was added to 300 mL of each fermentation medium (in quadruplicate) at optical density at 660 nm (OD 660 nm) of 0.2. The bacteria were grown at 30°C without agitation under oxygen-limited conditions. Cell growth was monitored by OD for 72 h and samples collected for proteomic study were immediately frozen in liquid nitrogen and stored at -80°C until analysis.
Quantification of sugars and fermentation products
Sucrose, glucose, fructose, ethanol, sorbitol were quantified in the fermentative liquid using high performance liquid chromatography (HPLC), Aminex HPX87H column, with 5,0 μM sulfuric acid as mobile phase, at a flow rate of 0.6 mL min-1 and temperature of 60°C. The mobile phase was prepared with ultrapure water and thereafter filtered on a 0.22 μm microporous membrane. Levan production was determined as previously reported (Viikari, 1984). The fermentation medium was centrifuged at 5000 xg for 10 min for biomass removal and the supernatant precipitated in 75% ethanol. The levan content was expressed in g/L by dry weight.
Extraction and quantification of cellular proteins
The stored cells were thawed on ice for 15 min, washed three times with PBS to remove culture medium residues and collected by centrifugation at 6000 g for 10 min. The precipitate was extracted by the Trizol method, following the instructions of manufacturer. This method was previously established as the most efficient for extracting Z. mobilis proteins (Cavalcanti and Malafaia, 2015). Protein quantification was performed using 2-D Quanti kit (GE Life Sciences) according to the manufacturer's recommendations. The BSA protein standard was used to obtain a calibration curve.
Two-dimensional gel electrophoresis (2-DE) and image analysis
Isoelectric focusing (IEF) was carried out in IPGphor 3 (GE Healthcare, Piscataway, NJ, USA) in pre-cast of 11 cm 1Q31718 dry strip and immobilized pH gradient of 3-10, which were hydrated with 500 μg of proteins dissolved into rehydration solution (7 M urea, 2 M thiourea, 2% CHAPS, 19.4 mM DTT, IPG buffer/pH 3-10 non-linear 1.0% (GE Life Sciences) and 0.005% bromophenol blue). IEF was performed at 20°C in a stepwise manner under conditions as follows: 100V for 2 hours, 250V for 2 hours, 500V for 2 hours, 1000V for 2 h, 8000V gradients for 4 h, and 8000V for a total of 66.000Vh. SDS-PAGE in the second dimension was performed with 12% gels using 16 mA/gel for 0.5 h, followed by 32 mA/gel at 16°C. Protein spots on gels were visualized by staining with the improved Coomassie Brilliant Blue G-250 method (Neuhoff, 1988). Each protein sample was analyzed by 2-DE at least three times. Protein gels were analyzed with ImageMaster 2D Platinum 6.0 software (GE Healthcare, Piscataway, NJ, USA) according to the protocols provided by the manufacturer. Spot detection was performed, followed by manual corrections to remove artifacts that did not correspond to proteins and the quantity of each site was normalized by the total intensity of valid spots. The normalized protein spots were compared and those that showed increase or decrease in abundance to more than double were considered differentially expressed, with values of p<0.05. Selected protein spots were subjected to gel digestion for identification by MALDI-TOF/TOF MS.
Identification of spots differentially expressed by mass spectrometry
Manually selected sites were excised from gels and digested with trypsin (Shevchenko and Shevchenko, 2001). Resultant peptides were extracted into solution containing 5% trifluoracetic acid and 50% acetonitrile, concentrated in Speed-Vac (Eppendorf) for 30 minutes and subjected to mass spectrometric analysis. MS and MS/MS spectra were obtained on the MALDI-TOF/TOF Autoflex III (Bruker Daltonics spectrometer) using α-cyano matrix. The MS in conjunction with MS/MS spectra were searched by the MASCOTE software (version 2.2, Matrix Science, London, UK) and the UniprotKB/Swiss-Prot database.
Results and Discussion
Growth of Z. mobilis ZAG-12 in media with different sucrose concentration
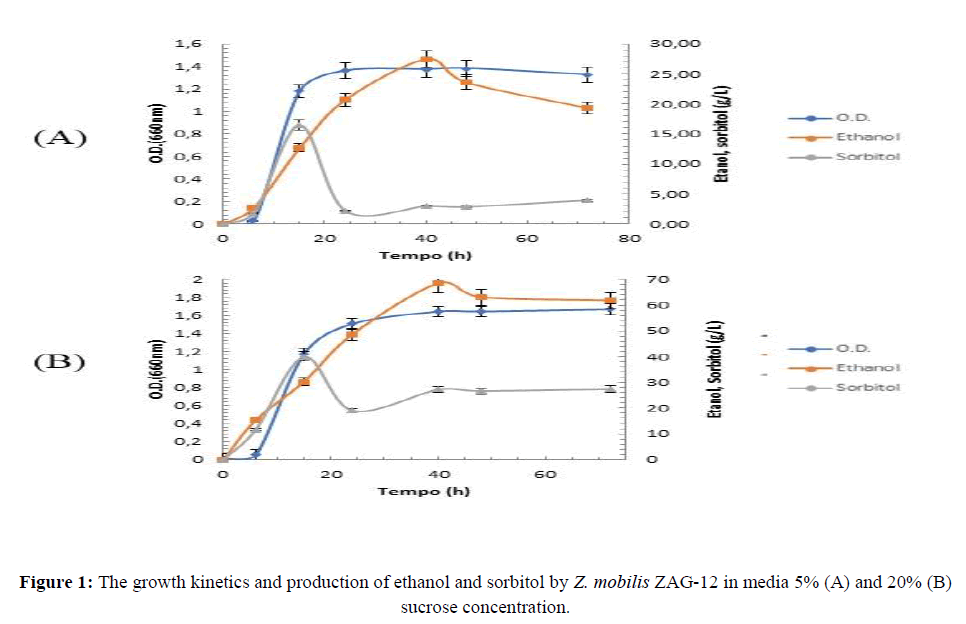
The growth kinetics of Z. mobilis ZAG-12 was spectrophotometrically measured (OD 660 nm) in two media containing different sucrose concentrations (50 g/L or 200 g/L) (Figure 1). An overview of fermentation performance of Z. mobilis ZAG-12 in media with different sucrose concentrations are shown in Table 1. Z. mobilis ZAG-12 was able to grow in both conditions studied, reaching a maximum value (OD 660 nm of 1.9) after 40 hours. The rates of specific growth were similar in both conditions, with values of 0.32 h-1 and 0.34 h-1 in media with 50 g/L sucrose and 200 g/L sucrose, respectively. The formation of a Lag phase was observed and lasted approximately 10 hours in both culture. After this interval, the exponential phase started, which is the period that the microorganism exhibits intense metabolic activity. The pH ranged from 6.5 (initial) to about 4.5 after 12 h of fermentation in both conditions, this value was maintained until the final period (72 h).
| Measured and calculated variables | Sucrose | |
|---|---|---|
| 5% | 20% | |
| Cells ( g/L) | 1.44 ± 0.03 | 1.81 ± 0.03 |
| Substrates consumed (%) | 88.38 ± 3.21 | 96.51 ± 2.16 |
| Ethanol ( g/L) | 19.32 ± 1.67 | 61.73 ± 2.78 |
| Biochemical efficiency - ethanol production (%) | 79.21 ± 6.84 | 61.32 ± 2.76 |
| Substrate utilization rate ( g/L/h) | 0.66 ± 0.02 | 2.6 ± 0.05 |
| Substrate conversion factor in ethanol (Yp/s - g/g) | 0.40 ± 0.03 | 0.31 ± 0.02 |
| Substrate conversion factor in cells (Yx/s - g/g) | 0.03 ± 0.002 | 0.009 ± 0.001 |
| Volumetric ethanol yield ( g/L/h) | 0.27 ± 0.03 | 0.86 ± 0.04 |
| Sorbitol ( g/L) | 3.97 ± 0.29 | 27.4 ± 1.49 |
Table 1: Fermentation performance of Z. mobilis ZAG-12 performance in media with different sucrose concentrations.
Substrate consumption in the culture media was high but not complete, the medium with 5% sucrose showed the lowest percentage of consumption (88.38%), while the medium with 20% sucrose presented 96.51% consumption. These values are higher than those showed in a study where the effects of high sucrose concentration on levan production by Z. mobilis CCT 4494 were evaluated. These authors found a substrate consumption rate of 84.4% and 81.3% within media containing 10% and 20% sucrose, respectively (Garcia-Cruz, 2009). Regarding the substrate breaking performance, ZAG-12 cultivation in medium containing 5% sucrose provided a faster utilization of this carbohydrate. After 6 hours of fermentation, only 6 g/L of residual sucrose was detected (approximately 11% degradation), whereas the medium with 20% sucrose had 167 g/L (only 16.5% degradation). These results suggest an inhibition of the enzymatic activity of the invertase due to substrate increased concentration. The decrease in sucrose hydrolysis efficiency due to high concentrations of sugars presented was previously suggested (Doelle and Greenfield, 1985).
An important feature for the use of Z. mobilis in biotechnological purposes is its reduced cellular growth in relation to yeasts. Z. mobilis usually allocate only 2% of the carbon source to produce biomass, while the most of the substrate is converted into ethanol, its main product (Yang and Fei, 2016). However, it is possible to observe an association of this growth with the production of ethanol as seen in Figure 1. The maximum ethanol production by ZAG-12 occurred in the medium with 20% sucrose in 40 hours of fermentation, obtaining 68.36 g/L (Figure 1B). In the other condition (5% sucrose), a total of 27.47 g/L was obtained (Figure 1C). These results are similar to those found for other strains such as Z. mobilis ATCC 29191 which produced about 54.88 g/L ethanol in a medium with 20% sucrose (Barros and Celligoi 2006). However, the best biochemical efficiency for ethanol production was observed in the medium with 5% sucrose with 79.21% of the maximum theoretical. The medium with 20% sucrose exhibited 61.32% of the maximum theoretical (Table 1). The volumetric productivity of ethanol at the end of fermentation was 0.27 g/L/h and 0.86 g/L/h in the media with 5% and 20% sucrose, respectively. Z. mobilis is well known for having biochemical efficiency close to the maximum theoretical in media containing only glucose. However, in media containing sucrose, lower efficiency is found, due to substrate deviation for the production of by-products such as levan and, mainly, sorbitol (Viikari, 1984).
Sorbitol production by Z. mobilis ZAG-12 was maximal in 15 hours of fermentation in both conditions. The best production (40.04 g/L) was observed in the medium with 20% sucrose (Figure 1A), whereas in 5% sucrose condition the sorbitol level was 16.47 g/L (Figure 1B). Similar studies have also shown that Z. mobilis strains are able to produced high levels of in media containing 20% sucrose, as shown for Z. mobilis ATCC 10988 (Lee and Huang, 1995) (24.3 g/L after 24 h of fermentation) and Z. mobilis ATCC 29191 (42.35 g/L after 36 h of fermentation) (Barros and Celligoi, 2006). All these data highlight the ability of Z. mobilis to produce sorbitol under osmotic pressure, suggesting an osmo-protective function. In fact, under different stress conditions (high temperatures, osmotic pressure and ethanol concentrations) (Lee and Huang, 1995), sorbitol production plays a key role in promoting cell growth and ethanol production (Sootsuwan and Thanonkeo, 2013). Sootsuwan in 2013 showed the importance of sorbitol under stress conditions such as temperature, ethanol concentration and osmotic pressure not only for cell growth but also for ethanol production. Thus, our results are in agreement with this evidence, since the highest concentrations of sugar and ethanol were found in the medium with 20% sucrose, where the highest production of sorbitol was also observed. In addition, the highest levan content was observed in the condition with 20% sucrose (7.38 g/L), while in the medium with 5% sucrose the levan concentration was almost 10 times lower (0.75 g/L).
Protein response of Z. mobilis grown in medium with high sugar concentration
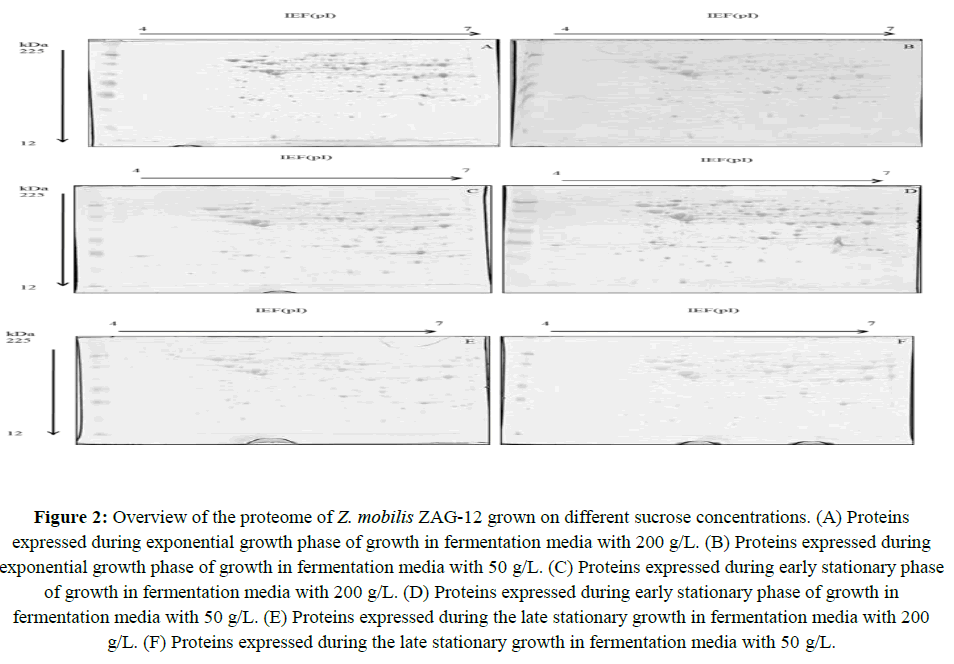
The 2D-gel proteomic approach was employed to identify differentially regulated proteins in Z. mobilis ZAG-12 cells in response to high sucrose concentration. Using gel-image software, about 352 spots were matched successfully on each gel. The 2D-E maps of Z. mobilis ZAG-12 in 200 g/L and 50 g/L conditions of sucrose are shown in Figure 2. Statistical analysis revealed that a total of 90 spots were differentially found in three fermentation period:
Figure 2: Overview of the proteome of Z. mobilis ZAG-12 grown on different sucrose concentrations. (A) Proteins expressed during exponential growth phase of growth in fermentation media with 200 g/L. (B) Proteins expressed during exponential growth phase of growth in fermentation media with 50 g/L. (C) Proteins expressed during early stationary phase of growth in fermentation media with 200 g/L. (D) Proteins expressed during early stationary phase of growth in fermentation media with 50 g/L. (E) Proteins expressed during the late stationary growth in fermentation media with 200 g/L. (F) Proteins expressed during the late stationary growth in fermentation media with 50 g/L.
(i) 28 spots in the exponential phase of growth (Table 2);
| Spot | Proteina | Microorganismb | Accession numberc | Scored | pIe | Nominal massf | Increased protein expression |
|---|---|---|---|---|---|---|---|
| B27 | 3-deoxy-7-phosphoheptulonate synthase | Z. mobilis ATCC 10988 | gi|384411581 | 247 | 5.02 | 49.35 | M20% |
| B28 | pyruvate kinase | Z. mobilis ZM4 | gi|56551048 | 80 | 6.26 | 51.52 | M20% |
| B29 | glyceraldehyde-3-phosphate dehydrogenase, type I | Z. mobilis ZM4 | gi|56551073 | 71 | 6.25 | 36.25 | M20% |
| B30 | 30S ribosomal protein S2 | Z. mobilis ZM4 | gi|56552052 | 100 | 5.89 | 27.42 | M20% |
| B31 | 50S ribosomal protein L9 | Z. mobilis ZM4 | gi|56552123 | 93 | 4.78 | 22.78 | M20% |
| B32 | 6-phosphoglucono-lactonase | Z. mobilis NCIMB 11163 | gi|260753946 | 132 | 5.18 | 25.54 | M20% |
| B33 | 6-phosphogluconate dehydrogenase | Gluconobacter oxydans H24 | gi|414341344 | 88 | 5.37 | 35.79 | M20% |
| B34 | pyruvate decarboxylase | Z. mobilis | gi|156563549 | 93 | 5.74 | 61.28 | M20% |
| B35 | pyruvate decarboxylase | Z. mobilis | gi|156563549 | 92 | 5.74 | 61.28 | M20% |
| B36*(MIX) | phosphomannomutase | Z. mobilis ATCC 10988 | gi|384411805 | 167 | 5.64 | 49.84 | M20% |
| B36*(MIX) | glucose-6-phosphate 1-dehydrogenase | Z. mobilis ATCC 29191 | gi|397676758 | 148 | 5.68 | 53.97 | M20% |
| B37 | aspartate 4-decarboxylase | Z. mobilis ATCC 29191 | gi|397677284 | 144 | 5.46 | 59.60 | M20% |
| B38 | arginyl-tRNA synthetase | Z. mobilis ZM4 | gi|283856343 | 237 | 5.43 | 63.36 | M20% |
| B39 | arginyl-tRNA synthetase | Z. mobilis ZM4 | gi|283856343 | 99 | 5.43 | 63.36 | M20% |
| B40 | trigger factor Tig | Z. mobilis ATCC 29191 | gi|397676242 | 186 | 4.92 | 55.21 | M20% |
| B41 | trigger factor Tig | Z. mobilis CP4 | gi|560136289 | 194 | 4.92 | 55.15 | M20% |
| B42 | phosphoserine phosphatase SerB | Z. mobilis ATCC 10988 | gi|384411128 | 146 | 4.88 | 32.26 | M20% |
| B45 | ketol-acid reductoisomerase | Z. mobilis ZM4 | gi|56552037 | 142 | 5.89 | 36.72 | M20% |
| B46 | phosphoglycerate kinase | Z. mobilis ZM4 | gi|56551074 | 151 | 5.97 | 41.47 | M20% |
| B47 | peptidase M1 membrane alanine aminopeptidase | Z. mobilis NCIMB 11163 | gi|260753680 | 225 | 5.84 | 96.26 | M20% |
| B49 | hypothetical protein ANA_C10416 | Anabaena sp. 90 | gi|414075712 | 93 | 4.97 | 22.93 | M5% |
| B51 | Zn-dependent alcohol dehydrogenase | Z. mobilis CP4 | gi|560136030 | 111 | 5.56 | 46.56 | M5% |
| B52 | 3-deoxy-7-phosphoheptulonate synthase | Z. mobilis ATCC 10988 | gi|384411581 | 201 | 5.02 | 49.35 | M5% |
| B53 | ROK family protein | Z. mobilis ZM4 | gi|56552615 | 170 | 5.79 | 32.93 | M5% |
aIdentification of the proteins by mass spectrometry.
bSpecies identified with the proteins encountered.
cAccession number in the database (nrNCBI).
dMASCOT score.
eIsoelectric point and
Ftheoretical molecular mass calculated from the equation Mr/PI of ExPASy.
Table 2: List of differentially expressed proteins in Z. mobilis ZAG-12 during the exponential growth phase in mediai 5% (M5%) or 20% (M20%) sucrose media.
(ii) 37 spots in the early stationary phase (Table 3); and
| Spot | Proteina | Microorganismb | Accession numberc | Scored | pIe | Nominal massf | Increased protein expression |
|---|---|---|---|---|---|---|---|
| B54 | 50S Ribosomal Protein L9 | Z. mobilis ZM4 | gi|56552123 | 90 | 4.78 | 22.78 | M5% |
| B55 | Cold-Shock DNA-Binding Domain-Containing Protein | Z. mobilis ZM4 | gi|283856223 | 143 | 5.57 | 26.49 | M5% |
| B56MIX | Ribosomal 5S Rrna E-Loop Binding Protein Ctc/L25/TL5 | Z. mobilis ZM4 | gi|56552806 | 157 | 5.6 | 23.36 | M5% |
| B56MIX | Cold-Shock DNA-Binding Domain-Containing Protein | Z. mobilis ZM4 | gi|283856223 | 79 | 5.57 | 26.49 | M5% |
| B57 | Fructokinase | Z. mobilis ATCC 29191 | gi|397677248 | 111 | 5.66 | 32.96 | M5% |
| B58 | Glucokinase | Z. mobilis ZM4 | gi|56551265 | 157 | 5.73 | 34.98 | M5% |
| B59 | 30S Ribosomal Protein S2 | Z. mobilis ZM4 | gi|56552052 | 125 | 5.89 | 27.42 | M5% |
| B60 | Cold-Shock DNA-Binding Domain-Containing Protein | Z. mobilis ZM4 | gi|283856223 | 103 | 5.57 | 26.49 | M5% |
| B61 | Ompa/Motb Domain-Containing Protein | Z. mobilis NCIMB 11163 | gi|260752248 | 141 | 7.03 | 38.57 | M5% |
| B62 | Class I And II Aminotransferase | Z. mobilis NCIMB 11163 | gi|260753133 | 145 | 6.1 | 43.99 | M5% |
| B63 | Hypothetical Protein | zeta proteobacterium SCGC AB-137-I08 | gi|517098087 | 100 | 5.95 | 48.79 | M5% |
| B64 | Phosphoglycerate Kinase | Z. mobilis ZM4 | gi|56551074 | 125 | 5.97 | 41.47 | M5% |
| B65 | Pyruvate Dehydrogenase (Acetyl-Transferring) E1 Component Subunit Alpha | Z. mobilis ZM4 | gi|56552502 | 190 | 6.01 | 38.80 | M5% |
| B66 | Ketol-Acid Reductoisomerase | Z. mobilis ZM4 | gi|56552037 | 171 | 5.89 | 36.72 | M5% |
| B67 | Zn-Dependent Alcohol Dehydrogenase | Z. mobilis CP4 | gi|560136030 | 155 | 5.56 | 46.56 | M5% |
| B68 | Deoxyhypusine Synthase-Like Protein | Z. mobilis ZM4 | gi|56551915 | 178 | 5.14 | 39.26 | M5% |
| B69 | Phosphoserine Phosphatase Serb | Z. mobilis ATCC 10988 | gi|384411128 | 160 | 4.88 | 32.26 | M5% |
| B70 | Cobalt Chelatase, Pcobs Small Subunit | Z. mobilis ZM4 | gi|56552440 | 134 | 4.83 | 37.05 | M5% |
| B71 | 3-Deoxy-7-Phosphoheptulonate Synthase | Z. mobilis ATCC 10988 | gi|384411581 | 200 | 5.02 | 49.35 | M5% |
| B72 | Peptidase M1 Membrane Alanine Aminopeptidase | Z. mobilis NCIMB 11163 | gi|260753680 | 225 | 5.84 | 96.26 | M5% |
| B73 | Thiamine Biosynthesis Protein Thic | Z. mobilis ATCC 10988 | gi|384411594 | 258 | 5.5 | 67.40 | M5% |
| B74 | Thiamine Biosynthesis Protein Thic | Z. mobilis ATCC 10988 | gi|384411594 | 278 | 5.5 | 67.40 | M5% |
| B75MIX | Phosphomannomutase | Z. mobilis ATCC 10988 | gi|384411805 | 207 | 5.64 | 49.84 | M5% |
| B75MIX | Glucose-6-Phosphate 1-Dehydrogenase | Z. mobilis ATCC 29191 | gi|397676758 | 112 | 5.68 | 53.97 | M5% |
| B76 | Glucose-6-Phosphate 1-Dehydrogenase | Z. mobilis ZM4 | gi|56551263 | 201 | 5.61 | 53.94 | M5% |
| B77 | Pyruvate Decarboxylase | Z. mobilis | gi|156563549 | 99 | 5.74 | 61.28 | M5% |
| B78 | Argininosuccinate Synthase | Z. mobilis ZM4 | gi|56551932 | 155 | 5.76 | 45.73 | M5% |
| B79 | Succinic Semialdehyde Dehydrogenase | Z. mobilis ZM4 | gi|4378160 | 132 | 5.79 | 49.75 | M5% |
| B80 | Elongation Factor G | Z. mobilis NCIMB 11163 | gi|260752977 | 224 | 4.99 | 76.86 | M5% |
| B81 | Pyruvate Kinase | Z. mobilis ZM4 | gi|56551048 | 154 | 6.26 | 51.52 | M20% |
| B82 | Pyruvate Kinase | Z. mobilis ZM4 | gi|56551048 | 159 | 6.26 | 51.52 | M20% |
| B83 | Glucan Biosynthesis Protein D | Z. mobilis ZM4 | gi|56551801 | 147 | 6.35 | 58.95 | M5% |
| B84 | Glyceraldehyde-3-Phosphate Dehydrogenase, Type I | Z. mobilis ZM4 | gi|56551073 | 189 | 6.25 | 36.25 | M20% |
| B85 | 2,3-Bisphosphoglycerate-Dependent Phosphoglycerate Mutase | Z. mobilis ATCC 29191 | gi|397675981 | 259 | 6.34 | 25.87 | M20% |
| B86 | 2,3-Bisphosphoglycerate-Dependent Phosphoglycerate Mutase | Z. mobilis ATCC 29191 | gi|397675981 | 215 | 6.34 | 25.87 | M20% |
| B87 | Superoxide Dismutase | Z. mobilis ZM4 | gi|56551956 | 124 | 6.36 | 23.06 | M20% |
| B88 | Superoxide Dismutase | Z. mobilis ZM4 | gi|56551956 | 149 | 6.36 | 23.06 | M20% |
| B89 | Cold-Shock DNA-Binding Domain-Containing Protein | Z. mobilis ZM4 | gi|283856223 | 88 | 5.57 | 26.49 | M5% |
| B90 | Carbonate Dehydratase | Z. mobilis ZM4 | gi|56552029 | 93 | 5.14 | 23.48 | M5% |
aIdentification of the proteins by mass spectrometry.
bSpecies identified with the proteins encountered.
cAccession number in the database (nrNCBI).
dMASCOT score.
eIsoelectric point and
Ftheoretical molecular mass calculated from the equation Mr/PI of ExPASy.
Table 3: List of differentially expressed proteins in Z. mobilis ZAG-12 during the early stationary growth phase in media with 5% (M5%) or 20% (M20%) sucrose media.
(iii) 25 spots in the late stationary phase (Table 4). Among these 90 spots, 78 (86.66%) were successfully identified after being submitted to mass spectrometry (MALDI-TOF), obtained their PMF and analyzed by MASCOT.
| Spot | Proteina | Microorganismb | Accession numberc | Scored | pIe | Nominal massf | Increased protein expression |
|---|---|---|---|---|---|---|---|
| B2 | Superoxide Dismutase | Z. mobilis ZM4 | gi|56551956 | 131 | 6.36 | 23.06 | M20% |
| B3 | Fructokinase | Z. mobilis ATCC 29191 | gi|397677248 | 89 | 5.66 | 32.96 | M5% |
| B4 | Glucokinase | Z. mobilis ZM4 | gi|56551265 | 135 | 5.73 | 34.98 | M5% |
| B5 | Cold-Shock DNA-Binding Domain-Containing Protein | Z. mobilis ZM4 | gi|283856223 | 128 | 5.57 | 26.49 | M5% |
| B6 | Ribosomal 5S Rrna E-Loop Binding Protein Ctc/L25/TL5 | Z. mobilis ZM4 | gi|56552806 | 93 | 5.6 | 23.36 | M5% |
| B8 | Phosphopyruvate Hydratase | Z. mobilis ZM4 | gi|56552504 | 148 | 4.99 | 45.88 | M20% |
| B11 | Phosphogluconate Dehydratase | Z. mobilis ZM4 | gi|11095426 | 90 | 6.56 | 63.17 | M5% |
| B12 | Peptidase M1 Membrane Alanine Aminopeptidase [ | Z. mobilis NCIMB 11163 | gi|260753680 | 94 | 5.84 | 96.26 | M20% |
| B14 | ATP-Dependent Chaperone Clpb | Z. mobilis NCIMB 11163 | gi|260753989 | 132 | 5.4 | 95.73 | M20% |
| B17(mix) | Phosphomannomutase | Z. mobilis ATCC 10988 | gi|384411805 | 156 | 5.64 | 49.84 | M20% |
| B17(MIX) | Glucose-6-Phosphate 1-Dehydrogenase | Z. mobilis ZM4 | gi|56551263 | 127 | 5.61 | 53.94 | M20% |
| B19 | Thiamine Biosynthesis Protein Thic | Z. mobilis ATCC 10988 | gi|384411594 | 191 | 5.50 | 67.40 | M5% |
| B20 | Pyruvate Decarboxylase | Z. mobilis | gi|156563549 | 71 | 5.74 | 61.28 | M5% |
| B21 | Pyruvate Decarboxylase | Z. mobilis | gi|156563549 | 107 | 5.74 | 61.28 | M5% |
| B22 | Deoxyhypusine Synthase-Like Protein | Z. mobilis ZM4 | gi|56551915 | 152 | 5.14 | 39.26 | M5% |
| B23 | Carbonate Dehydratase | Z. mobilis subsp. mobilis ZM4 | gi|56552029 | 108 | 5.14 | 23.48 | M5% |
| B24 | 6-Phosphogluconolactonase | Z. mobilis NCIMB 11163 | gi|260753946 | 155 | 5.18 | 25.54 | M5% |
| B25 | Alcohol Dehydrogenase | Z. mobilis ATCC 29191 | gi|397675985 | 160 | 5.70 | 36.57 | M5% |
| B26 | 3-Deoxy-7-Phosphoheptulonate Synthase | Z. mobilis ATCC 10988 | gi|384411581 | 209 | 5.02 | 49.35 | M5% |
aIdentification of the proteins by mass spectrometry.
bSpecies identified with the proteins encountered.
cAccession number in the database (nrNCBI).
dMASCOT score.
eIsoelectric point and
Ftheoretical molecular mass calculated from the equation Mr/PI of ExPASy.
Table 4: List of differentially expressed proteins in Z. mobilis ZAG-12 during the late stationary growth phase in media with 5% (M5%) or 20% (M20%) sucrose media.
These spots corresponded to 39 proteins, since in some cases the identification of adjacent spots indicated the same protein (Figure 3). This fact is attributed to the presence of isoforms due to post-translational or proteolytic degradation processes. During the exponential growth phase, twenty proteins were differentially expressed (16 were up-regulated and 4 down-regulated). In the early stationary phase, we found thirty-one proteins (five were up-regulated and twenty-six were down-regulated). Finally, in the late stationary phase we observed the lowest number of differentially expressed proteins (eighteen proteins: 6 up-regulated and 12 down-regulated).
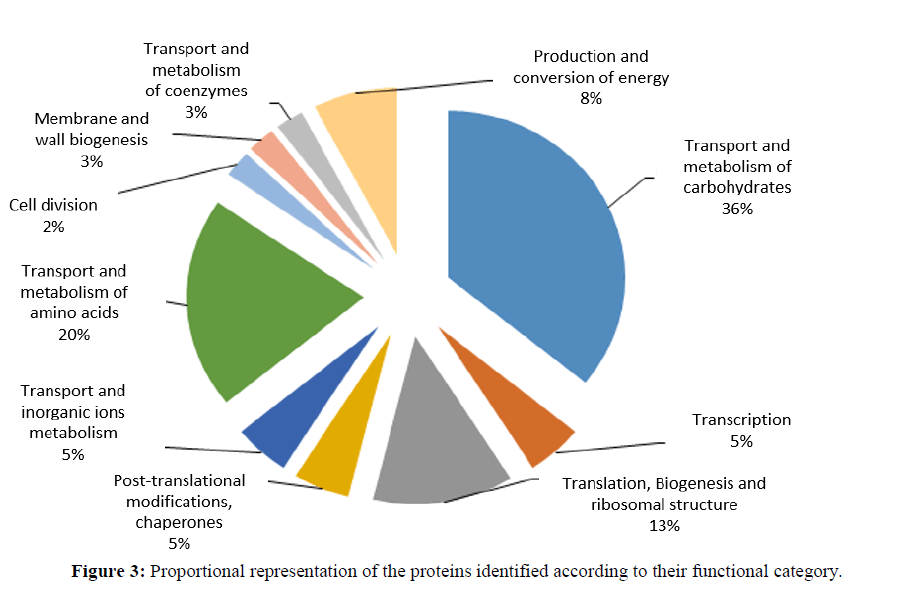
The differentially expressed proteins identified could be grouped in 10 functional categories: (i) Transport and metabolism of carbohydrates (36%); (ii) Transcription (5%); (iii) Translation, biogenesis and ribosomal structure (13%); (iv) Post-translational modifications and chaperones (5%); (v) Transport and metabolism of inorganic ions (5%); (vi) Transport and metabolism of amino acids (20%); (vii) cell division (2%); (viii) Membrane/cell wall biogenesis (3%); (ix) Transport and metabolism of coenzymes (3%); (x) Production and energy conversion (8%).
Our proteomic analysis indicates a global reorganization of carbohydrate metabolism, production and energy conversion of Z. mobilis ZAG-12 during fermentation with high sucrose concentration. The first change detected was an increase in glycolytic pathway (up regulation of kinase pyruvate and glyceraldehyde-3-phosphate dehydrogenase) in a medium with 20% sucrose. These data suggest a complete adaptation of the glycolytic pathway for the conversion of excess glucose/fructose to pyruvate. In addition, pyruvate decarboxylase was found to be up-regulated in medium with high sugar concentration. It is a key enzyme for ethanol and CO2 production as the final fermentation products of Z. mobilis (Pei and Erixon, 2010). Pyruvate decarboxylase is described in few bacterial genera as the enzyme that catalyzes the non-oxidative decarboxylation of pyruvate to produce acetaldehyde plus CO2. This enzyme also participates in the regeneration of NAD+. The regeneration of NAD+ exerts an advantageous influence on ethanolic fermentation due to alcohol dehydrogenase being NADH dependent (Hoppner and Doelle, 1983). Another important change was shown at the glycolysis control point in Z. mobilis, with upregulation of glucose-6-phosphate 1-dehydrogenase in medium with high sugar concentration (Snoep and Arfman, 1996).
Another protein found up-regulated in high sucrose condition was phosphomannomutase. This enzyme uses substrates of glycolytic pathway for synthesis glycoprotein and glycolipids of membrane, sugar nucleotides, exopolysaccharides (Huang and Li, 2013). Phosphomannomutase is also important for osmotic stabilization (Empadinhas and Marugg, 2001) and biofilm formation (Zimaro and Thomas, 2014). However, its role in Z. mobilis is still not well described. Interestingly, in our study, its expression was increased in the medium with high sugar content, a favorable condition for biofilm formation and increased cell surface hydrophobicity (Zikmanis and Shakirova, 2007) indicating a possible role of this enzyme in the structural response and modification of the bacteria surface.
Oxidative stress is defined as an interference in the balance between the production of reactive oxygen species (ROS), including free radicals, oxides, peroxides and the ability of biological systems to readily detect their presence and detoxify ROS or repair the resulting damage (Ortiz de Orue Lucana and Wedderhoff, 2012). In this study, the superoxide dismutase protein was identified with high expression in the early and late stationary phase. This antioxidant enzyme has previously been reported as responsible for the protection of the respiratory chain, reducing the formation of toxic products and increasing aerotolerance in Z. mobilis, therefore, the increase of antioxidant system allows remarkable improvements in the process of cultivation of Z. mobilis as shortening the lag phase and increasing ethanol production (Strazdina and Kravale, 2012; Balodite and Strazdina, 2014). Our results suggest that the increased level of superoxide dismutase could protect Z. mobilis from stress caused by the high concentration of sugar and ethanol by coordinating the level of ROS in an ideal balance.
During translation, elongation factors (EFS) are responsible for the delivery of aminoacyl-tRNA to the ribosome, and it has been proposed as a chaperone for unfolded and denatured proteins in E. coli (Yikilmaz and Chapman, 2014; Awai and Ichihashi, 2015). In our study, aminoacyl-tRNA was up-regulated in the exponential phase and along with elongation factors were down regulated in the early and late phases during growth in medium with high concentration of sucrose. In addition, the level of some ribosomal proteins (L9, S2) was also affected. Apart from protein biosynthesis, ribosomal proteins are also involved in DNA repair, cell death and transcriptional regulation (Siragusa and De Angelis, 2014). These results suggest that overexpression of protein-related translation and protein biosynthesis may be associated with initial adaptation to sugar concentration. The Tig trigger factor is a chaperone that participates in the de novo synthesis of proteins, with also cogitated as a grown factor. In Pseudomonas, is linked to growth and adaptation to stress conditions with heavy metals (Santos and Benndorf, 2004; Lee and Choi, 2009). We suggest that the higher biomass formation in the medium with 20% sucrose suffered from the tig trigger factor influence.
In the present work, we show that Z. mobilis ZAG-12 altered its protein expression and formation of products, such as ethanol and sorbitol, in response to a change in sucrose concentration. This response was characterized by proteomic analysis that detected 39 differentially expressed proteins, most related to transport and carbohydrate metabolism, amino acid metabolism and translation. The proteomic profile provided a better understanding of the physiological responses on how this promising microorganism redirects its metabolic flux to the production of ethanol and co-products.
About the Authors
Corresponding Author
L.A. Silva
Embrapa Agroenergia, Parque Estação Biológica, Brasília, DF, Brasil
- Email:
- proiupc_bog@unal.edu.co
References
- Ananthalakshmy VK and Gunasekaran P (1999) Isolation and characterization of mutants from levan-producing Zymomonas mobilis. J Biosci Bioeng 87: 214-217. https://doi.org/10.1016/s1389-1723(99)89015-x
- Awai T, Ichihashi N, Tetsuya Yomo (2015) Activities of 20 aminoacyl-tRNA synthetases expressed in a reconstituted translation system in Escherichia coli. Biochemistry and biophysics reports 3: 140-143. https://doi.org/10.1016/j.bbrep.2015.08.006
- Aziz MG, Michlmayr H, Kulbe KD, Hierro AMD et al. (2011) Biotransformation of pineapple juice sugars into dietetic derivatives by using a cell free oxidoreductase from Zymomonas mobilis together with commercial invertase. Enzyme Microb. Technol. 48: 85-91. https://doi.org/10.1016/j.enzmictec.2010.09.012
- Balodite E and Strazdina I (2014) Structure of the Zymomonas mobilis respiratory chain: oxygen affinity of electron transport and the role of cytochrome c peroxidase. Microbiol 160: 2045-2052. https://doi.org/10.1099/mic.0.081612-0
- Barros Md and Celligoi MAPC (2006) Synthesis of sorbitol by Zymomonas mobilis under high osmotic pressure. Braz J Microbiol 37: 324-328. https://doi.org/10.1590/s1517-83822006000300023
- Campos IA and Ximenes EA et al. (2013) Zymomonas mobilis culture protects against sepsis by modulating the inflammatory response, alleviating bacterial burden and suppressing splenocyte apoptosis. Eur J Pharm Sci 48: 1-8. https://doi.org/10.1016/j.ejps.2012.10.011
- Cavalcanti DR and Malafaia CB (2015) Protein extraction method for the proteomic study of Zymomonas mobilis during production of ethanol and levans. Genet Mol Res: GMR 14: 14406-14421. https://doi.org/10.4238/2015.november.18.4
- De Morais JO and Rios EM (1993) Zymomonas mobilis research in the Pernambuco Federal University. J. Biotechnol 31: 75-91. https://doi.org/10.1016/0168-1656(93)90138-d
- Doelle HW and Greenfield PF (1985) The production of ethanol from sucrose using Zymomonas mobilis. Appl. Microbiol. Biotechnol. 22: 405-410. https://doi.org/10.1007/bf00252781
- Einsfeldt K and Baptista IC (2016) Recombinant L-Asparaginase from Zymomonas mobilis: A Potential New Antileukemic Agent Produced in Escherichia coli. PloS one 11(6): e0156692. https://doi.org/10.1371/journal.pone.0156692
- Empadinhas N and Marugg JD (2001) Pathway for the synthesis of mannosylglycerate in the hyperthermophilic archaeon Pyrococcus horikoshii. J. Biol. Chem 276: 43580-43588. https://doi.org/10.1074/jbc.m108054200
- Erzinger GS and Vitolo M (2006) Zymomonas mobilis as catalyst for the biotechnological production of sorbitol and gluconic acid. Appl Biochem Biotechnol129-132: 787-794. https://doi.org/10.1385/abab:131:1:787
- Garcia-Cruz FMPGECH (2009) Zymomonas mobilis: A promising microrganism for alcoholic fermentation. Semina: Ciências Agrárias Agr Sci30: 361-380. https://doi.org/10.5433/1679-0359.2009v30n2p361
- Goderska K and Juzwa W (2015) Lactobionic acid production by glucose-fructose oxidoreductase from Zymomonas mobilis expressed in Escherichia coli. Biotechnol Lett 37: 2047-2053. https://doi.org/10.1007/s10529-015-1887-0
- Hoppner TC and Doelle HW (1983) Purification and kinetic characteristics of pyruvate decarboxylase and ethanol dehydrogenase from Zymomonas mobilis in relation to ethanol production. Eur J Appl Microbiol Biotechnol 17: 152-157. https://doi.org/10.1007/bf00505880
- Huang H and Li X (2013) Cloning, expression and characterization of a phosphoglucomutase/phosphomannomutase from sphingan-producing Sphingomonas sanxanigenens. Biotechnol Lett 35: 1265-1270. https://doi.org/10.1007/s10529-013-1193-7
- Kang HL and Kang HS (1998) A physical map of the genome of ethanol fermentative bacterium Zymomonas mobilis ZM4 and localization of genes on the map. Gene 206: 223-228. https://doi.org/10.1016/s0378-1119(97)00589-1
- Koti S and Govumoni SP (2016) Enhanced bioethanol production from wheat straw hemicellulose by mutant strains of pentose fermenting organisms Pichia stipitis and Candida shehatae. SpringerPlus 5: 1545. https://doi.org/10.1186/s40064-016-3222-1
- Kouvelis VN and Teshima H (2014) Finished Genome of Zymomonas mobilis subsp. mobilis Strain CP4, an Applied Ethanol Producer. Genome Announc 2. https://doi.org/10.1128/genomea.00845-13
- Lee K and Choi H (2009) Identification and expression of the tig gene coding for trigger factor from psychrophilic bacteria with no information of genome sequence available. Curr Microbiol 59: 160-166. https://doi.org/10.1007/s00284-009-9412-0
- Lee SM and Hong WK (2014) Enhancement of 1,3-propanediol production by expression of pyruvate decarboxylase and aldehyde dehydrogenase from Zymomonas mobilis in the acetolactate-synthase-deficient mutant of Klebsiella pneumonia. J Ind Microbiol Biotechnol 41: 1259-1266. https://doi.org/10.1007/s10295-014-1456-x
- Lee WC and Huang CT (1995) Enhancement of ethanol production from sucrose by Zymomonas mobilis by the addition of immobilized invertase. Enzyme Microb. Technol 17: 79-84. https://doi.org/10.1016/0141-0229(94)00042-p
- Liu C and Dong H (2010) Sorbitol production using recombinant Zymomonas mobilis strain. J Biotechnol 148: 105-112. https://doi.org/10.1016/j.jbiotec.2010.04.008
- Liu YF and Hsieh CW (2017) Effect of acetic acid on ethanol production by Zymomonas mobilis mutant strains through continuous adaptation. BMC biotechnol 17: 63. https://doi.org/10.1186/s12896-017-0385-y
- Lobato AV (2003) Produção de Oligossacarídeos por Zymomonas mobilis. Universidade federal de Pernambuco
- Mohagheghi A and Linger JG (2015) Improving a recombinant Zymomonas mobilis strain 8b through continuous adaptation on dilute acid pretreated corn stover hydrolysate. Biotechnology for biofuels 8: 55. https://doi.org/10.1186/s13068-015-0233-z
- Mohd Azhar SH and Abdulla R (2017) Yeasts in sustainable bioethanol production: A review. Biochem Biophys Rep 10: 52-61. https://doi.org/10.1016/j.bbrep.2017.03.003
- Morais Fd (1984/85) Spheroplasts of Zymomonas mobilis: production and regeneration. Revista do instituto de antibióticos 22: 157-163
- Ortiz de Orue Lucana D and Wedderhoff I (2012) ROS-mediated signalling in bacteria: Zinc-containing Cys-X-X-Cys redox centres and iron-based oxidative stress. J Signal Transduct 2012: 1-9. https://doi.org/10.1155/2012/605905
- Pei XY and Erixon KM (2010) Structural insights into the prereaction state of pyruvate decarboxylase from Zymomonas mobilis. Biochem 49: 1727-1736. https://doi.org/10.1021/bi901864j
- Pena-Castro JM and Del Moral S (2017) Biotechnological Strategies to Improve Plant Biomass Quality for Bioethanol Production. BioMed research international 2017: 7824076. https://doi.org/10.1155/2017/7824076
- Rostagno MA and Prado JM (2015) Subcritical and supercritical technology for the production of second generation bioethanol. Crit. Rev. Biotechnol 35: 302-312. https://doi.org/10.3109/07388551.2013.843155
- Lima SG (2011) Detection of bacteriocins in Zymomonas mobilis and RAPD fingerprinting of the producer strains.5: 19. https://doi.org/10.5897/ajpp11.328
- Santos PM and Benndorf D (2004) Insights into Pseudomonas putida KT2440 response to phenol-induced stress by quantitative proteomics. Proteomics 4: 2640-2652. https://doi.org/10.1002/pmic.200300793
- Shevchenko A and Shevchenko A (2001) Evaluation of the efficiency of in-gel digestion of proteins by peptide isotopic labeling and MALDI mass spectrometry. Anal. Biochem296: 279-283. https://doi.org/10.1006/abio.2001.5321
- Silbir S and Dagbagli S (2014) Levan production by Zymomonas mobilis in batch and continuous fermentation systems. Carbohydr Polym 99: 454-461. https://doi.org/10.1016/j.carbpol.2013.08.031
- Silva LC and Araujo JM (2015) Genetic variability analysis of Zymomonas mobilis strains from the UFPEDA microorganisms collection. Genetics and molecular research : GMR 14: 805-814. https://doi.org/10.4238/2015.february.2.5
- Siragusa S and De Angelis M (2014) Fermentation and proteome profiles of Lactobacillus plantarum strains during growth under food-like conditions. J Proteomics 96: 366-380. https://doi.org/10.1016/j.jprot.2013.11.003
- Snoep JL and Arfman N (1996) Control of glycolytic flux in Zymomonas mobilis by glucose 6-phosphate dehydrogenase activity. Biotechnol. Bioeng 51: 190-197. https://doi.org/10.1002/(sici)1097-0290(19960720)51:2%3C190::aid-bit8%3E3.3.co;2-l
- Sootsuwan K and Thanonkeo P (2013) Sorbitol required for cell growth and ethanol production by Zymomonas mobilis under heat, ethanol, and osmotic stresses. Biotechnol Biofuels 6: 180. https://doi.org/10.1186/1754-6834-6-180
- Strazdina I and Kravale Z (2012) Electron transport and oxidative stress in Zymomonas mobilis respiratory mutants. Arch Microbiol 194: 461-471. https://doi.org/10.1007/s00203-011-0785-7
- Tano MS and Buzato JB (2000) Sugar cane juice fermentation by Zymomonas mobilis CP4 subjected to inhibition by ethanol and high initial concentration of substrate. Braz Arch Biol Technol 43: 425-430. https://doi.org/10.1590/s1516-89132000000400012
- Vigants A and Upite D (2013) An influence of ethanol and temperature on products formation by different preparations of Zymomonas mobilis extracellular levansucrase. Folia microbiologica 58: 75-80. https://doi.org/10.1007/s12223-012-0185-7
- Viikari L (1984) Formation of levan and sorbitol from sucrose by Zymomonas mobilis. Appl Microbiol Biotechnol 19: 252-255. https://doi.org/10.1007/bf00251846
- Wang H and Cao S (2016) Very high gravity ethanol and fatty acid production of Zymomonas mobilis without amino acid and vitamin. J Ind Microbiol Biotechnol 43: 861-871. https://doi.org/10.1007/s10295-016-1761-7
- Yang S and Fei Q (2016) Zymomonas mobilis as a model system for production of biofuels and biochemicals. Microb Biotechnol 9: 699-717. https://doi.org/10.1111/1751-7915.12408
- Yang S and Franden MA (2014) Insights into acetate toxicity in Zymomonas mobilis 8b using different substrates. Biotechnol Biofuels 7: 140. https://doi.org/10.1186/s13068-014-0140-8
- Yang S and Mohagheghi A (2016) Metabolic engineering of Zymomonas mobilis for 2,3-butanediol production from lignocellulosic biomass sugars. Biotechnol Biofuels 9: 189.
- Yang S and Pan C2014) Elucidation of Zymomonas mobilis physiology and stress responses by quantitative proteomics and transcriptomics. Frontiers in microbiology 5: 246. https://doi.org/10.1186/s13068-016-0606-y
- Yang S and Pan C (2013) Systems biology analysis of Zymomonas mobilis ZM4 ethanol stress responses. PloS one 8: e68886.
- Yikilmaz E and Chapman SJ (2014) The interface between Escherichia coli elongation factor Tu and aminoacyl-tRNA. Biochem 53: 5710-5720. https://doi.org/10.1021/bi500533x
- Zhang X and Chen G (2009) Reduction of xylose to xylitol catalyzed by glucose-fructose oxidoreductase from Zymomonas mobilis. FEMS Microbiol Lett 293: 214-219. https://doi.org/10.1111/j.1574-6968.2009.01529.x
- Zikmanis P and Shakirova L (2007) Hydrophobicity of bacteria Zymomonas mobilis under varied environmental conditions. Process Biochem 42: 745-750. https://doi.org/10.1016/j.procbio.2007.01.002
- Zimaro T and Thomas L (2014) The type III protein secretion system contributes to Xanthomonas citri subsp. citri biofilm formation. BMC microbiol 14: 96. https://doi.org/10.1186/1471-2180-14-96
Keywords:
Download:
Full PDF- Share This