Identification of Clematidis radix et Rhizoma and its adulterants by core haplotype based on the ITS sequences
Received: February 27, 2018
Accepted: April 07, 2018
Published: April 15, 2018
Genet.Mol.Res. 17(2): gmr16039905
DOI: 10.4238/gmr16039905
Abstract
To develop a method to identify Clematidis radix et Rhizoma using sequence similarity and sequence-specific genetic polymorphisms based on the ITS sequences. DNA was extracted from leaves of Clematis mandshurica Rupr and C. hexapetala using a DNA extraction kit. ITS sequences were amplified by PCR, and analyzed in Contig Express, DNAman, and MEGA 5.0. The core haplotype was determined, and similarities between the core and other haplotypes were calculated. In total, 138 ITS sequences of C. mandshurica were obtained with a length of 611 bp. The similarity threshold between C. mandshurica and counterfeit species was 99%. Using specific mutation sites, we could identify C. chinensis, C. hexapetala, and C. mandshurica rapidly and accurately. A new DNA-based method has been established to rapidly and accurately identify Clematidis radix et Rhizoma.
Introduction
Clematidis radix et Rhizoma is the dry radix and rhizome of Clematis chinensis Osbeck, C. hexapetala Pall, and C. mandshurica, (Rupr Zhou Y, et al. 2012). In traditional Chinese medicine, Radix et Rhizoma Clematidis is applied to dispel wind dampness, flush the meridian, and treat rheumatic paralysis, limb numbness, and tendon spasm. Recent pharmacological studies have also shown that Clematidis radix et Rhizoma has anti-inflammatory, hypoglycemic, anti-hypertensive, and anti-tumor activities (Zhou Y, et al., 2012, Han W et al., 2013, Ionkova I 2011, Jung Up Park JN, et al. 2016, Mitjans M, et al. 2005), Yang J, et al. 2017). Because of wide application in clinics, adulterants of Clematidis radix et Rhizoma have proliferated in the market, including C. armandii Franch., C. finetiana H. Lév. et Vaniot, C. uncinata Champ., C. henryi Oliv., C. florida Thunb., C. chrysocoma Franch., C. lasiandra Maxim, C. peterae Hand.-Mazz., C. kerriana J.R. Drumm. et Craib, and C. leschenaultiana DC . Huang YY (2005), Song L et al. (2011), Li JS et al. (1980). These adulterants are traditionally identified based on the practitioner’s experience, variations in morphology due to the age of herbs and environmental conditions, and other factors. However, such factors are inadequate for accurate identification, because Clematidis radix et Rhizoma cannot be distinguished from adulterants morphologically or microscopically. However, genuine Clematidis and adulterants differ greatly in chemical composition and efficacy, and improper application may lead to drug safety issues. Song L, et al. (2011), Li JS et al., (1980), Guo LX et al., (2015). Therefore, it is of considerable importance to establish a rapid, accurate, and efficient method to identify genuine Clematidis.
DNA barcoding for species identification was first proposed by the Canadian zoologist Hebert in 2003. (Hebert PDN et al., 2003). This molecular diagnostic technology is based on short sequence fragments, and has been used since then to identify a variety of herbal medicines rapidly, efficiently, accurately, and objectively. (Yan HX et al., 2010, Hou DY et al., 2013, Chen JJ et al., 2015). Current methods of DNA barcoding include similarity searching, distance calculations, and tree-building. Similarity searching is mainly based on the BLAST algorithm. (Chen SL, et al., 2013, Cheng XL, et al., 2012 and Liu J, et al., 2011). However, due to the lack of a well-defined identification threshold, and frequently changing reference sequences, standardized identification is yet to be established for many herbal medicines. Consequently, a sample may be misidentified because a BLAST search may return two or more species with 99% similarity, or a neighbour-joining tree may cluster different species into a single branch. In addition, sequences from some species may not be present in existing databases, and are thus challenging to identify.
One way to identify a plant is to determine its haplotype, which typically consists of a DNA fragment with a number of nucleotide polymorphisms. The most frequent haplotype found within a species is considered the core haplotype, and all others are deemed rare. As would be expected, the similarity between the core and a rare haplotype of the same species is larger than the similarity between the core haplotype and a adulterant of the same genus. Therefore, a robust, well-defined similarity threshold value based on a large number of sequences could improve the accuracy, objectivity, and speed of DNA-based identification. In this study, we constructed a library of ITS sequences using 138 samples of C. mandshurica Rupr to determine the core haplotype, set the identification threshold value, and identify species-specific nucleotide variants, if any. Based on these parameters, we were able to identify samples of C. hexapetala, as well as C. chinensis and adulterant herbs from GenBank. Thus, we have established a rapid, accurate, and efficient molecular method to identify Clematidis radix et Rhizoma.
Materials and Methods
Plant materials
Leaves of C. mandshurica and C. hexapetala were collected from Liaoning Province, Heilongjiang Province, and Inner Mongolia Autonomous Region in China (Tables 1 and 2). ITS sequences from C. chinensis and adulterant herbs were downloaded from GenBank (Table 3).
| No. | Origin | Altitude (m) | Longitude | Latitude |
|---|---|---|---|---|
| 1 | Dalianzhuanghe, Liaoning Province | 43 | N31°41ʹ40ʺ | E123°2ʹ14ʺ |
| 2 | Fengcheng, Liaoning Province | 219 | N40°46ʹ 55ʺ | E123°54ʹ9ʺ |
| 3 | Hengren, Liaoning Province | 309 | N41°14ʹ15ʺ | E125°22ʹ12ʺ |
| 4 | Dengta, Liaoning Province | 173 | N41°22ʹ1ʺ | E123°31ʹ55ʺ |
| 5 | Kuandian, Liaoning Province | 310 | N40°58ʹ35ʺ | E125°0ʹ13ʺ |
| 6 | Benxi, Liaoning Province | 247 | N41°18ʹ0ʺ | E124°6ʹ25ʺ |
| 7 | Qingyuan, Fushun, Liaoning Province | 387 | N42°6ʹ58ʺ | E124°55ʹ39ʺ |
| 8 | Youyan, Anshan, Liaoning Province | 40 | N40°15ʹ51ʺ | E123°13ʹ41ʺ |
| 9 | Dandong, Liaoning Province | 204 | N39°1ʹ34ʺ | E129°15ʹ41ʺ |
| 10 | Qianshan, Liaoning Province | 129 | N41°1ʹ37ʺ | E123°8ʹ22ʺ |
| 11 | Kaiyuan, Liaoning Province | 98 | N42°22ʹ43ʺ | E124°0ʹ47ʺ |
| 12 | Tieling, Liaoning Province | 118 | N42°8ʹ38ʺ | E123°43ʹ53ʺ |
| 13 | Xifeng, Liaoning Province | 265 | N42°45ʹ26ʺ | E124°43ʹ21ʺ |
| 14 | Faku, Liaoning Province | 211 | N42°27ʹ59ʺ | E123°11ʹ59ʺ |
| 15 | Liaoyang, Liaoning Province | 248 | N42°56ʹ12ʺ | E123°22ʹ27ʺ |
| 16 | Xinbin, Liaoning Province | 258 | N41°47ʹ12ʺ | E124°38ʹ20ʺ |
| 17 | Fusong, Baishan, Jilin Province | 522 | N42°20ʹ35ʺ | E127°15ʹ48ʺ |
| 18 | Dongliao, Jilin Province | 423 | N42°48ʹ44ʺ | E124°57ʹ13ʺ |
| 19 | Lishu, Jilin Province | 255 | N46°0ʹ32ʺ | E130°40ʹ6ʺ |
| 20 | Yanji, Jilin Province | 160 | N42°55ʹ20ʺ | E129°35ʹ24ʺ |
| 21 | Liuhe, Jilin Province | 445 | N42°16ʹ8ʺ | E125°44ʹ40ʺ |
| 22 | Jiʹan, Tonghua, Jilin Province | 309 | N41°10ʹ40ʺ | E126°16ʹ10ʺ |
| 23 | Tonghe, Haʹerbin, Heilongjiang Province | 116 | N46°16ʹ53ʺ | E129°20ʹ43ʺ |
| 24 | Nancha, Yichun, Heilongjiang Province | 153 | N47°6ʹ47ʺ | E129°22ʹ20ʺ |
| 25 | Jiamusi, Heilongjiang Province | 102 | N46°45ʹ16ʺ | E130°22ʹ38ʺ |
| 26 | Tangyuan, Heilongjiang Province | 152 | N46°40ʹ1ʺ | E129°37ʹ32ʺ |
| 27 | Muleng, Mudanjiang, Heilongjiang Province | 297 | N44°53ʹ7ʺ | E130°33ʹ33ʺ |
| 28 | Hailin, Mudanjiang, Heilongjiang Province | 265 | N45°10ʹ49ʺ | E129°24ʹ10ʺ |
| 29 | Yilan, Heilongjiang Province | 109 | N46°20ʹ53ʺ | E129°34ʹ8ʺ |
| 30 | Shuangyashan, Heilongjiang Province | 450 | N46°22ʹ8ʺ | E131°6ʹ6ʺ |
Table 1. Voucher data for samples of Clematis mandshurica Rupr.
| No. | Origin | Altitude (m) | Longitude | Latitude |
|---|---|---|---|---|
| 1 | Nianzishan, Heilongjiang Province | 398 | N47°32ʹ44ʺ | E122°51ʹ17ʺ |
| 2 | Xiaoyingxiang, Yanji, Jilin Province | 160 | N42°55ʹ20ʺ | E129°35ʹ24ʺ |
| 3 | Dalianzhuanghe, Liaoning Province | 43 | N31°41ʹ40ʺ | E123°2ʹ13ʺ |
| 4 | Horqin Right Wing Front Banner, Inner Mongolia | 541 | N46°37ʹ15ʺ | E121°9ʹ24ʺ |
| 5 | Faku, Liaoning Province | 211 | N42°27ʹ59ʺ | E123°11ʹ59ʺ |
| 6 | Muleng, Heilongjiang Province | 297 | N44°53ʹ7ʺ | E130°33ʹ33ʺ |
Table 2. Voucher data for samples of C. hexapetala Pall.
| Var. | Accession numbers | |||
|---|---|---|---|---|
| Clematis chinensis | AB775161.1 | AB775169.1 | AB775162.1 | AB775170.1 |
| AB775163.1 | AB775171.1 | AB775164.1 | AB775172.1 | |
| AB775165.1 | AB775173.1 | AB775166.1 | AB775174.1 | |
| AB775167.1 | GU732584.1 | AB775168.1 | JF714641.1 | |
| Clematis armandii | FJ572047.1 | |||
| Clematis chrysocoma | GU732585.1 | GU732587.1 | GU732586.1 | |
| Clematis finetiana | GU732593.1 | JF714642.1 | ||
| Clematis florida | KC004031.1 | |||
| Clematis henryi | JF714645.1 | |||
| Clematis kirilowii | KC758681.1 | |||
| Clematis lasiandra | GU732600.1 | JF714640.1 | ||
| Clematis leschenaultiana | GU732603.1 | |||
| Clematis peterae | GU732614.1 | GU732615.1 | ||
| Clematis pogonandra | GU732618.1 | |||
| Clematis uncinata | GU732637.1 | JF714643.1 | ||
Table 3. GenBank accession numbers for ITS sequences of Clematis chinensis Osbeck and counterfeits of Clematidis radix et Rhizoma.
DNA extraction and PCR
Total DNA was extracted from approximately 0.1 g fresh leaves using a plant DNA extraction kit (Beijing Biomed Co., Ltd., Beijing, China), following the manufacturers protocol. Final DNA concentration was determined by spectrophotometry, and integrity was examined by electrophoresis on 1% (w/v) agarose. The universal primers ITS-F (5ʹ-AGAAGTCGTAACAAGGTTTCCGTAGG-3ʹ) and ITS-R (5ʹ-TCCTCCGCTTATTGATATGC-3ʹ) were used to amplify full length ITS sequences with the following program: Initial denaturation at 94°C for 5 min; 35 cycles at 94°C for 45 s, 55°C for 45 s, 72°C for 1 min; and final extension at 72°C for 10 min. Amplified fragments were sequenced by Sangon Biotechnology Co, Ltd. (Shanghai, China).
Sequence proof-reading and statistical analysis
Sequences were manually proofread in ContigExpress (Vector NTI, Invitrogen, Carlsbad, CA, USA), and aligned using blastn (available at BLAST NCBI, http://blast.ncbi.nlm.nih.gov/Blast.cgi). Species-specific nucleotide variants were identified with DNAman (Lynnon Biosoft, San Ramon, CA, USA) and MEGA-5.0. The core haplotype was determined, and similarities between the core and other haplotypes were calculated to set the identification threshold value.
Results
Amplification of ITS sequences
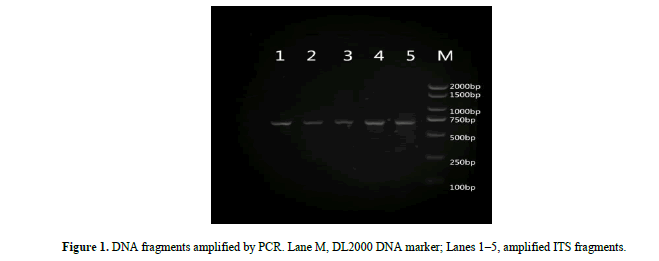
Electrophoresis confirmed that PCR products were of the expected length, highly pure and suitable for further analysis (Figure 1).
Sequence analysis
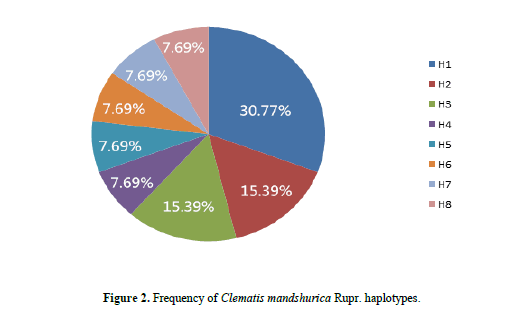
We obtained 138 ITS sequences of C. mandshurica Rupr from 21 locations. The length of the aligned matrix was 611 bp, with 50 polymorphic sites and 86 different haplotypes. After removing haplotypes due to hybridization, 8 distinct haplotypes were found. Polymorphic sites are listed in Table 4, and haplotype frequency is summarized in Figure 2.
| Haplotypes | Polymorphic sites (bp) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 5 | 6 | 19 | 26 | 55 | 94 | 150 | 428 | 566 | 591 | |
| H1 | A | C | C | A | A | A | C | G | C | C | C |
| H2 | * | - | - | * | * | * | T | * | * | T | G |
| H3 | * | * | * | * | * | * | T | * | * | T | G |
| H4 | - | - | - | - | - | * | * | * | * | T | G |
| H5 | * | * | * | * | * | * | T | * | * | * | G |
| H6 | * | * | * | * | * | T | * | * | * | * | G |
| H7 | * | * | * | * | * | T | T | A | T | T | G |
| H8 | * | * | * | * | * | * | * | * | * | T | G |
(Note: *indicates identity to H1; - indicates deletion).
Table 4. Polymorphic sites in Clematis mandshurica Rupr. haplotypes.
H1 was the most frequent haplotype at 30.77%, and was therefore considered the core haplotype for C. mandshurica Rupr. Accordingly, all other haplotypes were deemed rare. The similarity between the core and rare haplotypes was 99.35–100% based on BLAST alignment. On the other hand, the similarity between the core haplotype and the ITS fragment in C. hexapetala and C. chinensis was 97.23–98.85% and 97.46–98.73%, respectively. Furthermore, the similarity between the core haplotype and ITS sequences in adulterant herbs of the same genus was 93.07–97.86%. Therefore, 99% sequence similarity to the core haplotype was considered adequate to identify genuine C. mandshurica Rupr. DNA man inspection of ITS sequences in C. chinensis, C. hexapetala, C. mandshurica, and adulterants identified species-specific polymorphisms (Table 5). In particular, there was a clear distinction between adulterants and Radix et Rhizoma Clematidis at nucleotides 35, 86–89, 92, 159, 166, 422 and 555. In C. hexapetala, the bases at sites 89, 94, 95, 98, 158, 166, and 174 were G, G, A, T, G, T, and G, respectively, which significantly separated this species from C. chinensis and C. mandshurica. Finally, nucleotides 89, 98, and 570 were A, T, and T in C. mandshurica, but C, deletion, and C in C. chinensis, clearly separating the two species.
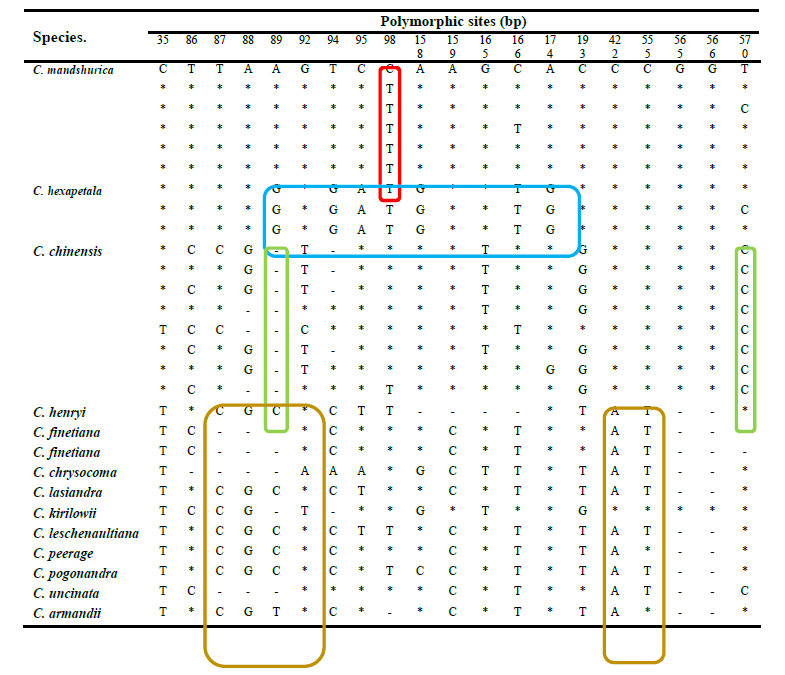
(Note : *indicates identity to the first sample; - indicates nucleotide deletion. Species-specific variants in C. mandshurica, C. hexapetala, C. chinensis, and counterfeits are highlighted in red, blue, green, and purple, respectively).
Table 5. Polymorphic sites in haplotypes of Clematis chinensis Osbeck, C. hexapetala Pall., C. mandshurica Rupr., and counterfeits of Clematidis Radix et Rhizoma
Discussion and Conculsion
Yang successfully used random amplified polymorphic DNA to assess the quality of Radix et Rhizoma Clematidis for 11 taxa of Clematis, but this method is poorly reproducible. On the other hand, PCR-single-strand conformation polymorphism was used to trace the origin of Clematis samples, but this technology is relatively cumbersome and time-consuming. Thus, ITS, ITS2, and psbA are commonly used as DNA barcodes for Radix et Rhizoma Clematidis. However, the accuracy, objectivity, and speed of identification based on these sequences are unsatisfactory, because comparison of a large number of sequences is required, along with an identification standard. In this study, we established a similarity threshold value for identifying C. mandshurica and adulterants. In addition, we found that Clematidis radix et Rhizoma could be identified accurately based on specific nucleotide sites in ITS sequences.
FINANCIAL SUPPORT AND SPONSORSHIP
National Science and Technology Major Project 2014X09304307001
Conflicts of interest
There are no conflicts of interest.
About the Authors
Corresponding Author
Chun-Sheng Liu
School of Chinese Pharmacy, Beijing University of Chinese Medicine, Beijing, China
- Email:
- max_liucs@263.net
References
- Pharmacopoeia Commission of PRC (2010) Pharmacopoeia of the people’s republic of China. Chemical Industry Press. Beijing 1: 234. https://doi.org/10.18356/94ce75d8-en
- Zhou Y, Guan Y, Shi J, Zhang X, et al. (2012) Development and validation of a chromatographic method for determining Clematichinenoside AR and related impurities . Chemistry Central Journal. 6(1): 150. https://doi.org/10.1186/1752-153x-6-150
- Han W, Xiong Y, Li Y, Fang W, et al. (2013) Anti-arthritic effects of clematichinenoside (AR-6) on PI3K/Akt signaling pathway and TNF-α associated with collagen-induced arthritis . Pharmaceutical Biology. 51(1): 13-22. https://doi.org/10.3109/13880209.2012.698287
- Ionkova I. (2011) Anticancer lignans from discovery to biotechnology. Mini Reviews in Medicinal Chemistry. 11(10): 843-856. https://doi.org/10.2174/138955711796575425
- Jung Up Park JN, Kim SJ, Na CS, Choi CH, et al. (2016) Chondroprotective and anti-inflammatory effects of ChondroT, a new complex herbal medication . Bmc Complementary and Alternative Medicine. 16(1): 1-11. https://doi.org/10.1186/s12906-016-1211-0
- Mitjans M, Vinardell MP (2005) Biological activity and health benefits of lignans and lignins . Trends in Comparative Biochemistry & Physiology,11: 55-62.
- Yang J, Song NL (2017) Chemical constituents from Clematidis radix et Rhizoma . Chinese Journal of Experimental Traditional Medical Formulae. 23(9): 41-45 [in Chinese].
- Huang YY (2005) Investigation on the original of Clematidis radix et Rhizoma . Guangdong Pharmaceutical Journal. 15(4): 4-6. [in Chinese]
- Song L, Zhang W, Zhang HM, Zhou XJ, et al. (2011) Investigation on the medicinal plant resources of Clematidis radix et Rhizoma in East China . Lishizhen Medicine and Materia Medica Research. 22(3): 725-727. [in Chinese].
- Li JS, Xiao PG, Lou ZC (1980) Research on botanical and pharmacognostical properties of Clematidis radix et Rhizoma . Acta Pharmaceutica Sinica. 15(5): 288-297. [in Chinese].
- Guo LX, Li R, Liu K, Yang J, et al. (2015) Structural characterization and discrimination of Chinese medicinal materials with multiple botanical origins based on metabolite profiling and chemometrics analysis: Clematidis radixet Rhizoma as a case study . Journal of Chromatography A. 1425: 129-140. https://doi.org/10.1016/j.chroma.2015.11.013
- Hebert PDN, Ratnasingham S, Dewaard JR (2003) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species . Proceedings. Biological Sciences / The Royal Society. 270 (S1): S96-S99. https://doi.org/10.1098/rsbl.2003.0025
- Hebert PDN, Cywinska A, Ball SL, Dewaard JR, et al (2003) Biological identifications through DNA barcodes . Proceedings. Biological Sciences / The Royal Society. 270(1512): 313-321. https://doi.org/10.5772/49967
- Yan HX, Yu J (2010) Research status of DNA-barcoding technology in plants . Chinese Bulletin of Botany. (1): 102-108. [in Chinese]
- Hou DY, Song JY, Yao H, et al (2013) Molecular identification of Corni Fructus and its counterfeits by ITS/ITS2 sequences . Chinese Journal of Natural Medicines. 11(2): 121-127. https://doi.org/10.3724/sp.j.1009.2013.00121
- Chen JJ, Zhao QS, Liu YL, Zha SH, et al. (2015) Identification of maca (Lepidium meyenii Walp.) and its adulterants by a DNA-barcoding approach based on the ITS sequence . Chinese Journal of Natural Medicines. 13(9): 653-659. https://doi.org/10.1016/s1875-5364(15)30062-5
- Chen SL, Yao H, Han JP, Xin TY, et al. (2013) Guiding principle of molecular identification of DNA-barcoding in Chinese herbal medicine . China Journal of Chinese Materia Medica. 38(2): 141-148. [in Chinese]
- Cheng XL, Liao CL, Liu CS, Bai JF, et al. (2012) Research on the counterfeit of Cercis chinensis Bunge—Schisandra sphenanthera Rehd. based on NCBI database using DNA-barcoding identification . Journal of Chinese Medicinal Materials. 37(17): 2534-2537. [in Chinese]
- Liu J, Liu CS, Wei SL, Zhang J, et al (2011) Analysis of rDNA-ITS sequences and similarity of radix paeoniae . Journal of Chinese Medicinal Materials. 34(10): 1517-1521. [in Chinese]
- Tamura K, Peterson D, Peterson N, Stecher G, et al (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods . Molecular Biology and Evolution. 28(10): 2731-2739. https://doi.org/10.1093/molbev/msr121
- Yang L (2005) Research on the standardization of quality specification of Clematidis radix et Rhizoma [D]. Chengdu University of Chinese Medicine. 2005. [in Chinese]
- An R, Chen ML, Wang XY, Zhang XQ, et al (2013) Identification of Clematis species by PCR-SSCP analysis . Chinese Pharmaceutical Journal. 48(17): 1247-1249.
- Zeng X, Li L, Ye N, Yao H, et al (2011) Identification of Clematidis radix et Rhizoma and its counterfeits based on ITS2 sequence . Global Traditional Chinese Medicine. 4(4): 264-269. [in Chinese]
- Feng SS, Zheng SH, Li YK, Huang LF. (2014) Identification of Clematidis radix et Rhizoma and its counterfeits using DNA-barcoding . Acta Pharmaceutica Sinica. 49 (2): 260-266. [in Chinese]
- Zheng AY. Research of Radix et Rhizoma Clematidis based on ITS2 sequences . Chinese and Foreign Medical Research, 2014, 12(8): 155-156.
Keywords:
Download:
Full PDF- Share This