Inexpensive metagenomic DNA extraction protocol with high quality from marine sediments contaminated by petroleum hydrocarbons
Received: June 01, 2017
Accepted: August 14, 2017
Published: September 21, 2017
Genet.Mol.Res. 16(3): gmr16039743
DOI: 10.4238/gmr16039743
Abstract
Marine environments are a reservoir of relevant information on dangerous contaminants such as hydrocarbons, as well as microbial communities with probable degradation skills. However, to access microbial diversity, it is necessary to obtain high-quality DNA. An inexpensive, reliable, and effective metagenomic DNA (mgDNA) extraction protocol from marine sediments contaminated with petroleum hydrocarbons was established in this study from modifications to Zhou’s protocol. The optimization included pretreatment of sediment with saline solutions for the removal of contaminants, a second precipitation and enzymatic degradation of RNA, followed by purification of mgDNA extracted by electroelution. The results obtained indicated that the modifications applied to 12 sediments with total petroleum hydrocarbon (TPH) concentrations from 22.6-174.3 (μg/g dry sediment) yielded 20.3-321.3 ng/μL mgDNA with A260/A280 and A260/A230 ratios of 1.75 ± 0.08 and 1.19 ± 0.22, respectively. The 16S rRNA amplification confirmed the purity of the mgDNA. The suitability of this mgDNA extraction protocol lies in the fact that all chemical solutions utilized are common in all molecular biology laboratories, and the use of dialysis membrane does not require any sophisticated or expensive equipment, only an electrophoretic chamber.
Introduction
Metagenomics is a culture-independent method (Cottrell et al., 2005) that efficiently captures the genome of a microbial community (Mineta and Gojobori, 2016). Marine environments are a reservoir of relevant information on dangerous contaminants such as hydrocarbons, as well as microbial communities with probable degradation skills. Moreover, the information generated can be used as a biological indicator for the characterization of aqueous pollution (Santhiya et al., 2011).
However, to access microbial diversity it is necessary to obtain high-quality DNA (Devi et al., 2015), an essential requirement for many of the next-generation high-throughput techniques (Fortin et al., 2004; Sharma et al., 2005). The construction of a metagenomic library represents the entire genetic complement of a single habitat, depending on the efficiency of the DNA extraction method (Felczykowska et al., 2015). In this regard, commercial kits have been used for purification of metagenomic DNA (mgDNA), but their uptake has been restricted due to the limited number of samples, their high cost, and importantly their lower quality and yields (Fortin et al., 2004) than laboratory protocols. The implementation of protocols is necessary in many cases, although most methods must be adapted according to the contaminants present in the sample. Therefore, the development of total DNA isolation protocols should start with the removal of contaminants before cell lysis (Fang et al., 2015). In particular, marine soils are commonly rich in fulvic and humic acids, which are frequently co-extracted with DNA (Nair et al., 2014) because they possess physical and chemical characteristics similar to DNA molecules and thus are too difficult to remove (Fitzpatrick et al., 2010). Heavy metals are also present in marine sediments and strongly inhibit any reaction in PCR amplification (Matheson et al., 2009). Contaminants other than proteins should be kept to a minimum (Felczykowska et al., 2015). In this regard, it has been demonstrated that pretreatment of sediment samples to remove inhibitors and contaminants can be crucial. The application of common but appropriate reagents, such as EDTA or Tween-20, promotes their elimination for retrieval of total DNA from contaminated sediments (Fang et al., 2015), although the inclusion of glass beads is frequently used for their elimination (Tanase et al., 2015). One technique for purifying DNA with remnant humic acids is the electrodialysis method, which consists of the application of an electrical field to the DNA isolate, employing a dialysis membrane for specific extraction of DNA from the solution. This method has been employed for successful DNA recovery from petroleum-polluted metallic environmental samples (Rodríguez-Mejía et al., 2008). This study describes successful modifications made to a DNA extraction protocol initially developed for soils and improved for sediments contaminated with hydrocarbons and collected in different seasons. The improvements include sediment pretreatment and DNA purification.
Materials and Methods
Marine sediment samples
A total of 12 soil samples were collected during the rainy and dry seasons from three sites of two coastal locations: Progreso and Sisal in Yucatan, Mexico. Samples were stored differentially at 4° and -20°C for further TPH and molecular genetic analysis respectively. The samples of rainy season were labeled PAEL, PBEL, and PCEL for samples from Progreso and SAEL, SBEL, SCEL for those from Sisal. Meanwhile, dry season samples were labeled PAES, PBES, PCES for samples from Progreso and SAES, SBES, and SCES for those from Sisal.
Sample preparation for petroleum-hydrocarbon extraction
The sediments were processed to determine moisture percentage as suggested Fernández et al. (2006). Twenty grams of sediment was dried at 28°C for 48 h and then pulverized into fine particles to obtain a higher surface area in contact with organic solvents. The moisture percentage was obtained by differences in weight before and after calcination at 80°C for 24 h.
Hydrocarbon extraction by mechanical shaking
This method is efficient and inexpensive regarding time and the solvents used. It was developed from US EPA methods 3500B and 3540C, as suggested by Schwab et al. (1999) and Arce-Ortega et al. (2004). The sediment (5 g) was mixed with 3 g anhydrous Na2SO4 and vortexed until homogeneous. Then, 5 mL dichloromethane was added, and it was homogenized for 45 s. Subsequently, the supernatant was recovered after centrifugation (4°C, 4072 RCF for 10 min) and the precipitate was resuspended. This was executed until 15 mL organic extract was obtained, which was brought to constant weight for its quantification.
Gravimetric quantification
This method was based on 821-B94-004 (EPA method) and quantified TPH in the samples by their weight balance. Firstly, 15 mL organic extract was placed in glass vials at a constant weight and vaporized under nitrogen flux until concentrated to dryness. Then, the weight differences corresponding to TPH content were recorded, taking into account the moisture measured for the calculation.
Molecular genetic analysis
Pretreatment of collected samples
The treatment for humic and fulvic acid removal was executed according to Fang et al. (2015) using different quantities (0.65, 0.45, and 0.25 g) of sediment sample PCEL and 3 mL saline buffer with 0.1 M EDTA, 0.1 M Tris, pH 8.0, 1.5 M NaCl, 0.1 M NaH2PO4 and Na2HPO4. Subsequently, samples were incubated at 65°C for 15 min at 180 rpm. Finally, they were centrifuged at 25°C for 6 min at 9979 RCF. The supernatant was discarded, and the pellet was resuspended in 3 mL saline buffer. This was repeated 3 times until a clear supernatant was obtained.
Metagenomic DNA extraction
Metagenomic DNA was extracted from all 12 soil samples by the method of Osborn et al. (2000) modified from Zhou et al. (1996). Fresh sediment (0.25 g) was suspended in 600 μL extraction buffer [100 mM Tris, 100 mM EDTA, 100 mM sodium phosphate buffer, pH 7, 5 μL proteinase K (V302B, Promega) (10 mg/mL), 9 μL lysozyme (LDB0308, Biotech) (100 mg/mL)] and incubated at 37°C for 30 min at 150 rpm. Subsequently, 150 μL 10% SDS, 225 μL 5 M NaCl and 75 μL 5% CTAB/1.5 M NaCl were added, and samples were incubated at 65°C for 15 min. The sample was then submitted to freeze-thaw using liquid nitrogen for 5 min and thawed at 65°C for 10 min. This step was repeated twice. After centrifugation at 4°C for 10 min at 1995 RCF (Digicen-21R, Orto Alresa), the supernatant was transferred, and the pellet was suspended by vortexing for 10 s in 200 μL extraction buffer and 50 μL 10% SDS. It was then incubated at 65°C for 10 min. After centrifugation at 4°C for 10 min at 1995 RCF, the supernatant was removed and pooled with that obtained from the first centrifugation. An equal volume of phenol:chloroform:isoamyl alcohol (25:24:1) was added to the pooled supernatant and mixed, and then centrifuged at 4°C for 5 min at 1995 RCF. The upper layer was transferred to a new tube to add an equal volume of chloroform:isoamyl alcohol (24:1), and it was centrifuged again at 4°C for 5 min at 1995 RCF. The upper layer (aqueous phase) was transferred to a new tube with 0.7 v isopropanol (at -20°C), and 0.3 v 3 M sodium acetate was added. After mixing by inversion, it was centrifuged at 4°C for 30 min at 13,709 RCF. The supernatant was then discarded, and the pellet was suspended in 75 μL 70% ethanol, and subjected to centrifugation at 4°C for 10 min at 9520 RCF. The supernatant was discarded, and the pellet was left to dry. Subsequently, the pellet was suspended in 400 μL TE and incubated at 65°C for 30 min. It was then transferred to 1.5-mL tubes and centrifuged at 4°C for 20 min at 21,420 RCF. The supernatant was recovered and transferred to a new tube to add 5 μL RNase (Ribonuclease A from bovine pancreas, R6513, Sigma) (10 mg/mL), and then was subjected to 37°C for 30 min. For mgDNA precipitation, 0.7 v isopropanol at -20°C and 0.6 v 3 M sodium acetate were added, mixed by inversion and centrifuged at 21,420 RCF for 15 min. The supernatant was discarded, and 200 μL 70% ethanol (at -20°C) was added to the pellet and centrifuged at 4°C for 10 min at 9520 RCF. The supernatant was discarded, and the pellet was suspended in 25 μL water. The DNA extraction protocol modified from Zhou et al. (1996) was applied to isolates of sulfate-reducing bacteria from cultures enriched in Postgate B medium (Postgate, 1984). From 20 mL bacterial culture, 1 mL culture and 1 mL TE buffer (10 mM Tris-HCl and 1 mM EDTA) were centrifuged (21,420 RCF for 15 min at 4°C). Then, the supernatant was discarded, and the precipitate was resuspended in 1 mL bacterial culture and 1 mL TE buffer. This procedure was executed until the medium was exhausted. DNA extraction was applied using 1 mL extraction buffer, 42 μL proteinase K (10 mg/mL), and 75 μL lysozyme (100 mg/mL). This solution was mixed by inversion and incubated at 37°C for 30 min, and then 250 μL 10% SDS, 374 μL 5 M NaCl and 125 μL 5% CTAB/1.5 M NaCl were added. This solution was incubated at 65°C for 15 min and centrifuged (21,420 RCF for 10 min at 4°C). The supernatant was divided into two tubes to add a volume of phenol:chloroform:isoamyl alcohol (25:24:1), mixed by inversion and centrifuged (5355 RCF for 5 min at 4°C). The supernatant was mixed with 0.7 v isopropanol (at -20°C) and 0.3 v 3 M sodium acetate. This was mixed by inversion and centrifuged (21,420 RCF for 30 min at 4°C). The precipitate was washed using 70% ethanol and resuspended in 400 μL TE buffer (10 mM Tris-HCl, 1 mM EDTA) for incubation at 65°C for 30 min in a water bath. It was then centrifuged (21,420 RCF for 20 min at 4°C), the supernatant was recovered, 5 μL RNase A (10 mg/mL) was added, and it was incubated at 37°C for 30 min. The genomic DNA was precipitated at room temperature with 280 μL isopropanol (at -20°C) and 120 μL 3 M sodium acetate and centrifuged (21,420 RCF for 15 min at 4°C). The supernatant was discarded, and the pellet was washed with 70% ethanol and dried to resuspend in 25 μL double-distilled water.
Metagenomic DNA yield and purity estimates
The nucleic extract (5 μL) and Orange G (03756-25G, Sigma) (2 μL) were loaded on agarose gel at 1% with 1X TAE and 1.2 μL Gelred (41003, Biotium). The sizes of fragments were referenced to 1-kb DNA ladder (Invitrogen). The bands were visualized by Photo Documentation (1302 GDS, Labnet). Metagenomic DNA was checked for purity and concentration by Nanodrop (2000, ThermoScientific) on absorbance read at 230, 260, and 280 nm.
Metagenomic DNA purification by dialysis membrane
The purification of mgDNA was executed as follows. Fragments of mgDNA from 1% agarose gel were frozen at -20°C for 30 min. Meanwhile, 7 cm dialysis membrane (D9777, Sigma) was prepared by rinsing in double-distilled water for a few minutes and then in 1X TAE buffer. A dialysis tube was prepared with a rubber band tightened at the end of the membrane. Then, the frozen fragments and 400 μL 1X TAE were introduced and a rubber band placed at the other end. The dialysis tube was placed inside an electrophoresis chamber at 100 mV for 15 min. Then, the 1X TAE buffer was removed and transferred to a new tube with 200 μL 70% ethanol (at -20°C) and centrifuged at 4°C for 10 min at 9520 RCF. The precipitate was suspended in 20 μL double-distilled water and stored at -20°C until use.
Amplification of 16S rRNA
The 16S rRNA gene amplification by PCR was executed obtaining a fragment of 484 bp using specific primers for the V1-V4 coding region: 28F (5'-GAGTTTGATCNTGGCTCAG-3') and 534R (5'-ATTCCGCGGCTGCTGG-3'). The PCR for 10 μL was executed with 1X PCR buffer, 0.16 mM dNTPs, 1.5 mM MgCl2, 0.5 μM of each primer, 0.02 μL of Taq polymerase recombinant (11615-010, Invitrogen) and 10 ng mgDNA. The cyclic profile was initial denaturalization at 94.5°C for 4 min, 25 cycles of 94.5°C for 1 min, 61.9°C for 1 min and 72°C for 1 min, with a final elongation at 72°C for 10 min (T100-ThermalCycler, Bio-Rad). DNA extracted from pure culture was used as a positive amplification control.
Results and Discussion
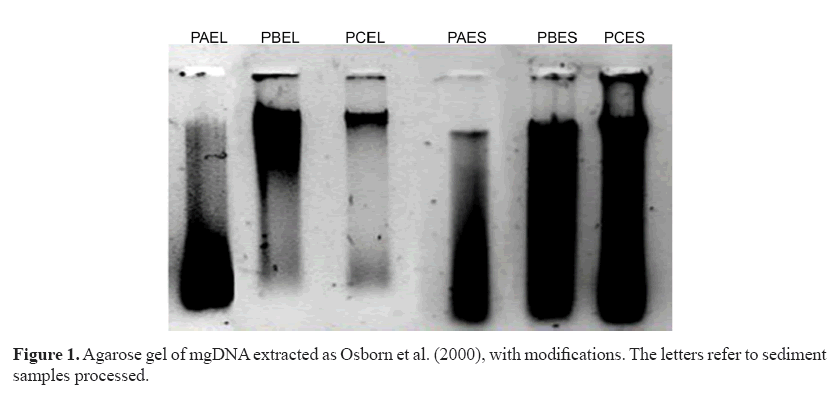
Numerous protocols have been used and optimized for soil samples (Steffan et al., 1988; Zhou et al., 1996; Osborn et al., 2000), and commercial kits are available for DNA extraction from soils contaminated with polycyclic aromatic hydrocarbons (PAHs) (Mahmoudi et al., 2011). However, most offer low DNA yields, compromising bacterial diversity. An inexpensive, reliable, and effective mgDNA extraction protocol from marine sediments contaminated with petroleum hydrocarbons was established in this study. This method, developed originally by Zhou et al. (1996) for a variety of sediment and soil types and further modified by Osborn et al. (2000), was developed and optimized in this study. The sediment samples contained higher TPH concentrations in rainy than in dry seasons (Table 1). In fact, the TPH concentrations were at the top of the maximum range permitted by UNESCO (70 μg/g) for coastal sediments in the Gulf of Mexico. Higher concentrations in rainy than in dry seasons have been documented for PAHs, with runoff comprising the major PAH source in the rainy season in Azuabie Creek in the Niger Delta (Daka and Ugbomeh, 2013). High PAH concentrations were also measured in sediments during the rainy season in an estuary in Sinaloa, Mexico, where contamination is closely related to human activities, such as domestic and industrial discharge, automobile exhausts, and street runoff (Jaward et al., 2012). In the area where the samples were taken for this study, the cause was probably runoff of contaminants from anthropogenic (maritime ports, fisheries, industrial, and urban) activities performed in the area (Valenzuela et al., 2005). It is important to highlight that this is the first study since that of Valenzuela et al., in 2005, and Mexico does not have any official measure on maximum permissible limits to marine sediments. The protocol of Osborn et al. (2000) developed for mgDNA extraction was modified by RNase enzyme addition and a second isopropanol precipitation followed by ethanol washes. The modifications produced higher mgDNA yields of 41.9-914.5 ng/μL (Table 2) than those of the Zhou et al. (1996) protocol with 2.5-26.9 μg/g. However, this modified protocol produced a higher A260/A280 ratio than the protocol of Zhou et al. (A260/280 of 1.35).
| Samples | HTP concentration (µg/g de dry sediment) |
| PAEL | 146.6 |
| PBEL | 174.3 |
| PCEL | 151.7 |
| SAEL | 48.9 |
| SBEL | 109.1 |
| SCEL | 81.9 |
| SAES | 22.6 |
| SBES | 104.0 |
| SCES | 73.4 |
| PAES | 88.1 |
| PBES | 104.2 |
| PCES | 85.2 |
Table 1: Concentrations of HTP measured in each sediment sample.
| Samples | mgDNA concentration (ng/µL) | ng/g of soil | 260/280 ratio | 260/230 ratio |
| PAEL | 914.5 | 365.8 | 1.8 | 2.41 |
| PBEL | 914.5 | 365.8 | 1.86 | 1.14 |
| PCEL | 98.1 | 39.24 | 1.74 | 0.18 |
| PAES | 463.2 | 185.28 | 1.81 | 1.16 |
| PBES | 116.8 | 46.72 | 1.76 | 1.02 |
| PCES | 41.9 | 16.76 | 1.83 | 1.19 |
Table 2: The mgDNA concentration from Osborn et al. protocol and it absorbance relations.
The extracts were visibly degraded and apparently contained RNA (Figure 1) despite increases in concentration (data not shown). The results obtained suggested the presence of remnants of contaminants that were co-extracted with mgDNA causing its degradation. To eliminate them, sediment pretreatment was performed before cell lysis. Different sample sizes of marine sediment (PCEL sample) were pretreated with saline solution as suggested by Fang et al. (2015).
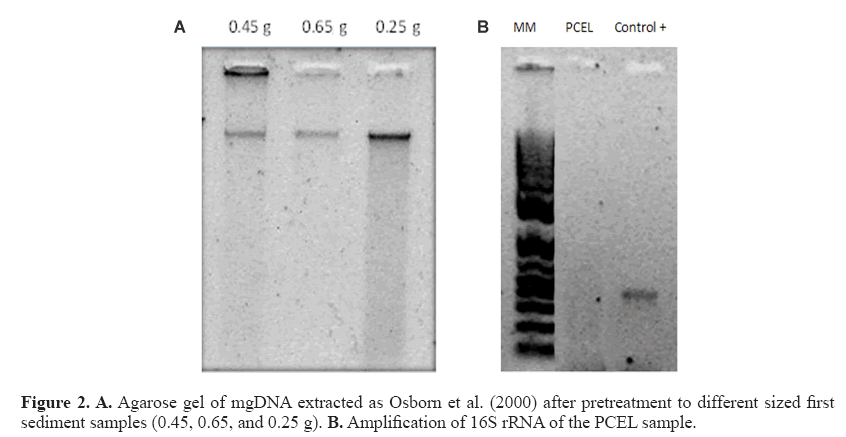
The mgDNA obtained after pretreatment was 265-772.5 ng/μL with substantially improved quality (Figure 2 and Table 3) because both the A260/A280 and A260/A230 ratios were higher than those obtained through Osborn’s and Zhou’s protocols. As suggested by the results obtained, higher sediment samples produce higher mgDNA yields.
| Sample sizes (g) | mgDNA concentration (ng/µL) | A260/280 nm | A260/230 nm |
| 0.65 | 772.5 | 1.74 | 1.63 |
| 0.45 | 272.5 | 1.83 | 1.63 |
| 0.25 | 265.0 | 1.81 | 1.71 |
Table 3: The mgDNA concentration and quality obtained from pre-treatment of different sizes of the sample PCEL.
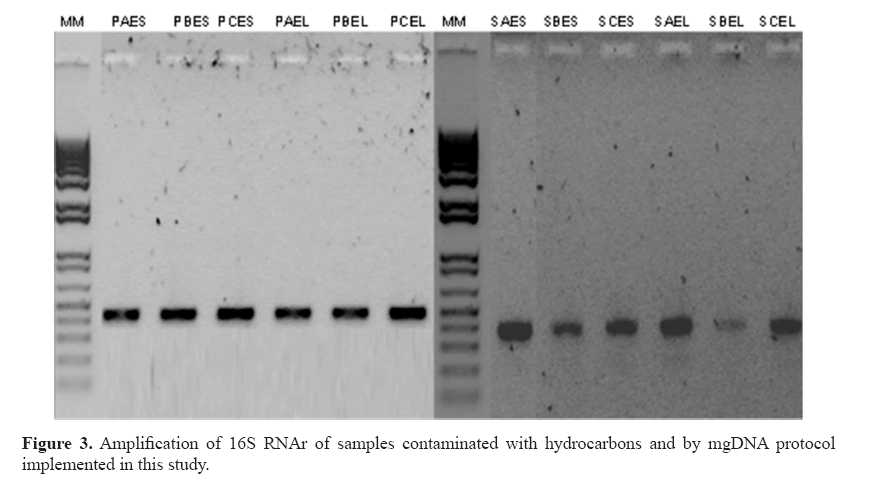
For practical purposes, 0.25 sediment was selected and executed in all samples with yields 161.8-389.6 ng/μL (Table 4). The chemicals comprising the saline solution (EDTA, Tris, NaCl, and phosphate buffer) were suitable. EDTA chelates metal ions and toxic materials (Klucakova, 2012), which leads to inactivation of lysozymes and proteinase K, and accelerates the dissolution of humic materials that bind metal ions or heavy metals (Lahiri and Schnabel, 1993; Klucakova, 2012). The mgDNA integrity was visualized by agarose electrophoresis with no RNA presence. However, amplification of the 16S RNA gene was not possible (Figure 2). The failures to obtain amplification results serve as confirmation of the presence of an inhibitory compound in the DNA sample, because the positive control was amplified (Cone et al., 1992). Humic substances or metals probably remained, inhibiting the amplification reaction. In particular, humic substances chelate Mg2+ ions during PCR, and therefore, indirectly affect the activity of Taq DNA polymerase (Tsai and Olson, 1992). Meanwhile, studies have indicated that co-purified metal ions inhibit DNA amplification (Combs et al., 2015), including calcium, which is also inhibitory. Metal ions can form soluble, high-molecular-weight complexes with humic substances in soils. Additionally, they can form adducts with DNA and can form crosslinks between DNA and proteins (Matheson et al., 2009). These chemical interactions can impair access to the DNA template during PCR. Dialysis membrane has been used with electroelution for isolation of DNA from oxidized metallic environments contaminated with petroleum (Rodríguez-Mejía et al., 2008). The procedure performed by using dialysis tubing facilitates the removal or exchange of small molecules from macromolecules in solution based on differential diffusion. The quality was adequate, following purification with a dialysis membrane, although yields were diminished lightly in some samples (Table 5). In fact, those samples with major loss of yield are because purification process was applied many days before to mgDNA extraction with pretreatment. The protocol for mgDNA extraction and purification was applied to all samples, indicating the reproducibility of this protocol that produces mgDNA of high quality and yield (Table 5), in comparison with Zhou’s or Osborn’s protocols. As a result, the mgDNA amplification results were successful (Figure 3). This result indicated that the modifications made to this protocol were highly suitable for samples highly contaminated by petroleum hydrocarbons.
| Samples name | mgDNA (ng/µL) | A260/A280 | A260/ A230 |
| PAES | 302.4 | 1.87 | 1.98 |
| PBES | 288.2 | 1.86 | 1.99 |
| PCES | 161.8 | 1.67 | 2.09 |
| PAEL | 359.9 | 1.88 | 1.90 |
| PBEL | 318.0 | 1.87 | 1.97 |
| PCEL | 273.3 | 1.88 | 1.98 |
| SAES | 239.3 | 1.81 | 1.92 |
| SBES | 247.5 | 1.83 | 1.98 |
| SCES | 251.1 | 1.87 | 2.01 |
| SAEL | 311.3 | 1.85 | 1.98 |
| SBEL | 309.8 | 1.81 | 2.09 |
| SCEL | 389.6 | 1.86 | 1.99 |
Table 4: Yield and quality of mgDNA from the protocol implemented with pre-treatment of sediment without purification for membrane of dialyses.
| Samples name | mgDNA (ng/µL) | A260/A280 | A260/A230 |
| PAES | 256.4 | 1.78 | 1.03 |
| PBES | 203.1 | 1.74 | 1.18 |
| PCES | 91.8 | 1.57 | 1.38 |
| PAEL | 321.3 | 1.74 | 1.08 |
| PBEL | 302.9 | 1.85 | 1.53 |
| PCEL | 21.9 | 1.83 | 1.07 |
| SAES | 25.8 | 1.85 | 1.03 |
| SBES | 21.9 | 1.71 | 1.09 |
| SCES | 51.85 | 1.67 | 1.69 |
| SAEL | 20.3 | 1.76 | 0.98 |
| SBEL | 42.2 | 1.80 | 1.23 |
| SCEL | 34.7 | 1.76 | 0.99 |
Table 5: Yield and quality of mgDNA from the new protocol implemented with pre-treatment of sediment and purification with dialysis membrane on electroelution.
The suitability of this mgDNA extraction protocol lies in the fact that all chemical solutions utilized are common in all molecular biology laboratories, and the use of dialysis membrane does not require any sophisticated or expensive equipment, only an electrophoretic chamber. Additionally, it can be implemented for successful results from any soils contaminated with humic acids or hydrocarbons. In this protocol, the humic acids were eliminated by pretreatment of sediments with saline solutions. The inhibitory effects of amplification were eliminated with dialysis tubing with electroelution. The adaptation of the mgDNA protocol with two noninvasive techniques may include additional advantages for providing a representative sampling of the metagenome population. The purity of the mgDNA obtained by this mgDNA protocol was further substantiated by the simple construction of a metagenomic library (in the process). The statistically significant improvement in the efficiency of DNA extraction invariably contributes to downstream manipulations by effectively reducing contaminants.
This report describes a DNA extraction method from sediments highly contaminated with hydrocarbons. DNA yields and quality were suitable for amplification regardless of the TPH concentration contained in the sediment samples. DNA was extracted from each sample, confirming its reproducibility. It is important to highlight that the chemical solutions used in this protocol are prevalent in molecular biology laboratories. Therefore, its execution does not require any special method and any laboratory can perform the protocol implemented here.
Acknowledgments
The authors gratefully acknowledge the financial support granted by the Consejo Nacional de Ciencia y Tecnología (CONACyT, Mexico, through project #166371) and postgraduate grants for I. García-Bautist (#394801). We acknowledge Paola Marfil Lara for the edition of figures.
About the Authors
Corresponding Author
R. Valdez-Ojeda
Unidad de Energía Renovable, Centro de Investigación Científica de Yucatán, Chuburná de Hidalgo, Mérida, Yucatán, Mexico
- Email:
- ruby.valdez@cicy.mx
References
- Arce-Ortega JM, Rojas-Avelizapa NG and Rodríguez-Vázquez R (2004). Identification of recalcitrant hydrocarbons present in a drilling waste-polluted soil. J Environ Sci Health A Tox Hazard Subst Environ Eng 39: 1535-1545. https://doi.org/10.1081/ESE-120037852
- Combs LG, Warren JE, Huynh V, Castaneda J, et al. (2015). The effects of metal ion PCR inhibitors on results obtained with the Quantifiler(®) Human DNA Quantification Kit. Forensic Sci. Int. Genet. 19: 180-189. https://doi.org/10.1016/j. fsigen.2015.06.013
- Cone RW, Hobson AC and Huang MLW (1992). Coamplified positive control detects inhibition of polymerase chain reactions. J. Clin. Microbiol. 30: 3185-3189.
- Cottrell MT, Waidner LA, Yu L and Kirchman DL (2005). Bacterial diversity of metagenomic and PCR libraries from the Delaware River. Environ. Microbiol. 7: 1883-1895. https://doi.org/10.1111/j.1462-2920.2005.00762.x
- Daka ER and Ugbomeh AP (2013). Polycyclic aromatic hydrocarbons in sediment and tissues of the crab callinectes pallidus from the Azuabie Creek of the upper bonny estuary in the Niger Delta. Res. J. Appl. Sci. Eng. Technol. 6: 2594-2600.
- Devi SG, Fathima AA, Radha S, Arunraj R, et al. (2015). Rapid and economical method for efficient DNA extraction from diverse soils suitable for metagenomic applications. PLoS One 10: e0132441. https://doi.org/10.1371/journal. pone.0132441
- Fang Y, Xu M, Chen X, Sun G, et al. (2015). Modified pretreatment method for total microbial DNA extraction from contaminated river sediment. FESE 9: 444-452.
- Felczykowska A, Krajewska A, Zieli√?¬?ska S and √?¬Āo√?¬? JM (2015). Sampling, metadata and DNA extraction - important steps in metagenomic studies. Acta Biochim. Pol. 62: 151-160. https://doi.org/10.18388/abp.2014_916
- Fitzpatrick KA, Kersh GJ and Massung RF (2010). Practical method for extraction of PCR-quality DNA from environmental soil samples. Appl. Environ. Microbiol. 76: 4571-4573. https://doi.org/10.1128/AEM.02825-09
- Fortin N, Beaumier D, Lee K and Greer CW (2004). Soil washing improves the recovery of total community DNA from polluted and high organic content sediments. J. Microbiol. Methods 56: 181-191. https://doi.org/10.1016/j. mimet.2003.10.006
- Fernández LC, Rojas NG, Roldán TG, Ramirez IME, et al. (2006). Manual de técnicas de análisis de suelos aplicadas a la remediación de sitios contaminados. Instituto Mexicano del Petróleo. Secretaria de Medio Ambiente y Recursos Naturales, Instituto Nacional de Ecología, México, D.F.
- Jaward FM, Alegria HA, Galindo RJG and Hoare A (2012). Levels of PAHs in the waters, sediments, and shrimps of Estero de Urias, an estuary in Mexico, and their toxicological effects. Thescientificworldjo 687034.
- Klucakova M (2012). Comparative study of binding behaviour of Cu(II) with humic acid and simple organic compounds by ultrasound spectrometry. TOCOLLSJ 5: 5-12. https://doi.org/10.2174/1876530001205010005
- Lahiri DK and Schnabel B (1993). DNA isolation by a rapid method from human blood samples: effects of MgCl2, EDTA, storage time, and temperature on DNA yield and quality. Biochem. Genet. 31: 321-328. https://doi.org/10.1007/ BF00553174
- Mahmoudi N, Slater GF and Fulthorpe RR (2011). Comparison of commercial DNA extraction kits for isolation and purification of bacterial and eukaryotic DNA from PAH-contaminated soils. Can. J. Microbiol. 57: 623-628. https:// doi.org/10.1139/w11-049
- Mineta K and Gojobori T (2016). Databases of the marine metagenomics. Gene 576: 724-728. https://doi.org/10.1016/j. gene.2015.10.035
- Matheson CD, Marion TE, Hayter S, Esau N, et al. (2009). Technical note: removal of metal ion inhibition encountered during DNA extraction and amplification of copper-preserved archaeological bone using size exclusion chromatography. Am. J. Phys. Anthropol. 140: 384-391. https://doi.org/10.1002/ajpa.21106
- Nair HP, Vincent H and Bhat SG (2014). Evaluation of five in situ lysis protocols for PCR amenable metagenomic DNA from mangrove soils. Biotechnol. Rep. (Amst.) 4: 134-138. https://doi.org/10.1016/j.btre.2014.09.008
- Osborn AM, Moore ERB and Timmis KN (2000). An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2: 39-50. https://doi.org/10.1046/j.1462-2920.2000.00081.x
- Postgate JR (1984). The Sulphate-Reducing Bacteria. Cambridge University Press, New York.
- Rodríguez-Mejía JL, Martínez-Anaya C, Folch-Mallol JL and Dantán-González E (2008). A two-step electrodialysis method for DNA purification from polluted metallic environmental samples. Electrophoresis 29: 3239-3244. https:// doi.org/10.1002/elps.200700829
- Sharma R, Ranjan R, Kapardar RK and Grover A (2005). Unculturable bacterial diversity: An untapped resource. Curr. Sci. 89: 72-77.
- Santhiya G, Lakshumanan C, Selvin J and Asha D (2011). Microbiological analysis of seawater and sediments in urban shorelines: Occurrence of heavy metals resistance bacteria on Chennai beaches, Bay of Bengal. Microchem. J. 99: 197-202. https://doi.org/10.1016/j.microc.2011.05.004
- Schwab AP, Su J, Wetzel S, Pekarek S, et al. (1999). Extraction of petroleum hydrocarbons from soil by mechanical shaking. Environ. Sci. Technol. 33: 1940-1945. https://doi.org/10.1021/es9809758
- Steffan RJ, Goksøyr J, Bej AK and Atlas RM (1988). Recovery of DNA from soils and sediments. Appl. Environ. Microbiol. 54: 2908-2915.
- Tanase A-M, Mereuta I, Chiciudean I, Ionescu R, et al. (2015). Comparison of total dna extraction methods for microbial community form polluted soil. Agric. Agric. Sci. Procedia. 6: 616-622. https://doi.org/10.1016/j.aaspro.2015.08.102
- Tsai YL and Olson BH (1992). Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl. Environ. Microbiol. 58: 2292-2295.
- Valenzuela SIS, Gold-Bouchot G and Ceja MV (2005). Hidrocarburos en Agua y Sedimentos de la Laguna de Chelem y Puerto Progreso, Yucatán, México. In: Golfo de México Contaminación e Impacto Ambiental: Diagnóstico y Tendencias (Botello AV, Rendon von Osten J, Gold-Bouchot G and Agraz-Hernández C, eds.). Universidad Autónoma de Campeche, Universidad Nacional Autónoma de México, Instituto Nacional de Ecología, 311.
- Zhou J, Bruns MA and Tiedje JM (1996). DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62: 316-322.
Keywords:
Download:
Full PDF- Share This