Inheritance of bacterial spot resistance in Capsicum annuum var. annuum
Published: April 20, 2017
Genet.Mol.Res. 16(2): gmr16029631
DOI: 10.4238/gmr16029631
Abstract
Since 2008, Brazil is the largest consumer of agrochemicals, which increases production costs and risks of agricultural products, environment, and farmers’ contamination. Sweet pepper, which is one of the main consumed vegetables in the country, is on top of the list of the most sprayed crops. The bacterial spot, caused by Xanthomonas spp, is one of the most damaging diseases of pepper crops. Genetic resistant consists of a suitable way of disease control, but development of durable resistant cultivars as well as understanding of plant-bacterium interaction is being a challenge for plant breeders and pathologists worldwide. Inheritance of disease resistance is often variable, depending on genetic background of the parents. The knowledge of the genetic base controlling such resistance is the first step in a breeding program aiming to develop new genotypes, bringing together resistance and other superior agronomic traits. This study reports the genetic basis of bacterial spot resistance in Capsicum annuum var. annuum using mean generation analysis from crosses between accessions UENF 2285 (susceptible) and UENF 1381 (resistant). The plants of each generation were grown in a greenhouse and leaflets were inoculated with bacterial strain ENA 4135 at 105 CFU/mL in 1.0 cm2 of the mesophyll. Evaluations were performed using a scoring scale whose grades ranged from 1.0 (resistant) to 5.0 (susceptible), depending on symptom manifestation. Genetic control of bacterial spot has a quantitative aspect, with higher additive effect. The quantitative analysis showed that five genes were the minimum number controlling bacterial spot resistance. Additive effect was higher (6.06) than dominant (3.31) and explained 86.36% of total variation.
Introduction
Sweet and chili peppers (Capsicum spp) are widely grown in several parts worldwide, being used as raw material in food, pharmaceutical, cosmetic, and most recently in the ornamental market (Rêgo et al., 2011). In Brazil, both are crops with socio-economic importance because they are strongly associated with family agriculture, generating job and income due to high demand for labor (Sudré et al., 2010).
Several factors are limiting for the cultivation of these plant species, such as diseases caused by different pathogens, which compromise not only yields but also fruit quality (Rêgo et al., 2011). Bacterial spot, caused by different Xanthomonas species, is one of the most destructive diseases affecting sweet and chili pepper crops (Potnis et al., 2015), causing great losses under high humidity conditions, intense rainfalls, and temperatures between 20° and 30°C (Kurozawa and Pavan, 2005). The bacterial spot causes plant defoliation, hindering its development, reducing photosynthesis and compromising total yield, and also exposes fruit to the sunlight leading to fruit depreciation, reducing its marketability.
Since 2008, Brazil is the largest pesticide consumer in the world (Pedlowski et al., 2012). In the specific case of sweet peppers, a report published by the Brazilian Public Health Association (ABRASCO) showed that this crop has the highest contamination index, wherein 91.8% of the samples contained pesticide residues, followed by strawberries (63.4%) and cucumber (57.4%) (Associação Brasileira de Saúde Coletiva Dossiê - ABRASCO, 2015).
Genetic resistance to diseases is an effective mechanism that aims to minimize environmental and food contaminations by continuous spraying of pesticides (Riva-Souza et al., 2009). Genetic control of bacterial spot resistance in sweet peppers has been the focus of studies seeking for resistance sources and resistant cultivars abroad and in Brazil (Cook and Stall, 1982; Jones et al., 2002; Riva et al., 2004; Riva-Souza et al., 2007; Stall et al., 2009; Vallejos et al. 2010; Moreira et al., 2015).
Five dominant (Bs1, Bs2, Bs3, Bs4, and Bs7) and two recessive genes (bs5 and bs6) controlling bacterial spot resistance have been identified (Stall et al., 2009; Potnis et al., 2012). Also, Riva et al. (2004) reported that at least three recessive genes control the resistance to X. euvesicatoria in C. annuum. However, bacterial spot resistance conferred by dominant genes was not durable, since of each gene described can be overcome by specific races of the pathogen in field. Quantitative resistance should be emphasized as a more valuable alternative to control bacterial spot in Capsicum plants (Stall et al., 2009).
In a plant breeding program, holding reliable information on the genetic basis of a particular trait helps to define the most appropriate breeding strategy to be adopted (Cruz et al., 2014). This study reports the genetic basis of bacterial spot resistance by crossing susceptible and resistant accessions of C. annuum var. annuum. The minimum number of genes controlling resistance and other genetic parameters were estimated to generate subsidies for further development of Capsicum-resistant cultivars.
Material and Methods
Genotypes and generations
Generations F1, F2, BC1, and BC2 originated from crossing between two genotypes of C. annuum var. annuum, identified as UENF 2285 (female parent) and UENF 1381 (male parent) (Figure 1), both from the UENF germplasm bank. UENF 2285 is a variety of sweet pepper with squared fruit, which is susceptible to bacterial spot. UENF 1381 is a chili pepper (pungent) that has been used as a source of resistance to bacterial spot in the Capsicum-breeding program developed by UENF.
Seedlings of all generations were produced at the UENF, in Campos dos Goytacazes, RJ, Brazil (21°19'23"S latitude and 41°19'40"W longitude). This stage was carried out from December 2014 to April 2016. Seedlings were sown in 128-cell polystyrene trays, remaining in a growth chamber at 28°C. After the seedlings reached four to five leaves, they were transplanted to 500-mL pots containing a mixture of soil, sand, and manure (1: 1: 1, volume ratio). Hereafter, plants were left in greenhouse, and crop handling followed recommendations by Filgueira (2012), with some adaptations for this environment.
The crossings were performed early in the morning or late afternoon when buds were at the pre-anthesis. Female parent buds were emasculated and identified with a wool cloth. For pollen extraction from male parents, flowers were collected in the morning and dried under fluorescent bulbs. Afterward, pollen was removed and transferred to a gelatin capsule, being stored inside amber bottles with silica gel, in a refrigerator at ±5ºC, for later manual pollination. Emasculated flowers were pollinated and covered with paper bags to avoid further contamination.
Eighty crossings between UENF 2285 x UENF 1381 were carried out resulting in 24 hybrid fruit. Backcrosses were performed using 117 artificial crosses between UENF 2285 x F1 (BC1) and 66 between UENF 1381 x F1 (BC2). Ten plants of each parent and from hybrids were used, resulting in 101 fruits from BC1 and 37 from BC2. For F2 generation, 253 self-fertilizations were made in F1 generation, providing 11 fruits (Figure 2).
Bacterial spot inoculation and resistance assessment
Twenty plants of P1, P2, and F1, plus 200 plants of F2 and 40 plants of each backcross were used to evaluate resistance to bacterial spot. The bacterial strain ENA 4135, which was characterized by Riva et al. (2004) based on the differentiating genotypes proposed by Jones et al. (1998), was inoculated. The water-preserved strain (Castellani, 1939) was recovered in DYGS (Rodrigues Neto et al., 1986) liquid medium under agitation, for 36 h at 28°C. Thereafter, bacterial suspensions were transferred with Drigalsky’s loop to Petri dishes containing solid DYGS medium. After 36 h in a bacteriological incubator (28°C), bacterial colonies were suspended in sterile water and cell concentrations adjusted to 108 CFU/mL, at 600 nm and an absorbance of 0.300 (Aguiar et al., 2000). Such concentration was applied for qualitative analysis to evaluate the hypersensitivity reaction (HR). Then, suspension (108 CFU/mL) was subjected to serial dilution in distilled water to reach a concentration of 105 CFU/mL, for quantitative resistance evaluation.
Inoculation was performed 38 days after transplanting, in two leaves of the plant upper middle third, by infiltration of a bacterial suspension with the above-mentioned concentrations, in 1.0 cm2 of the mesophyll (Riva et al., 2004). HR was assessed 24 and 48 h after inoculation; it was solely considered the presence or the absence of an immediate and drastic cellular response of plants in contact with the pathogen.
Reaction to bacterial spot was quantitatively ascertained by means of a score scale. The scores consisted of: 1 - no visible symptoms, 2 - spotted chlorosis, 3 - yellowish leaves with some necrotic spots, 4 - necrotic spots, and 5 - total necrosis (Figure 3). This evaluation started 5 days after inoculation and lasted for 7 days. At the end, scores below 2 were classified as resistant plants, and those above 2 were classified as susceptible (Riva-Souza et al., 2009).
Original rating was used to calculate the area under disease progress curve (AUDPC) as proposed by Shaner and Finney (1977):
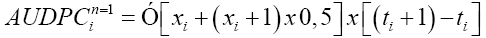 (Equation 1)
(Equation 1)
where n is the number of assessments; xi is the disease incidence or severity;
Analysis of variables
A quantitative approach was used to assess the results. This analysis was based on AUDPC analysis means and variances of the parental generations (F1, F2, BC1, and BC2). Each generation means were analyzed according to additive-dominant model, wherein averages varied only due to homozygosis (m), additive effect (a), and dominance deviation (d). Genetic parameters were estimated by weighted least squares.
From each generation variance analysis, the following estimates were obtained for the AUDPC results (Cruz, 2013):

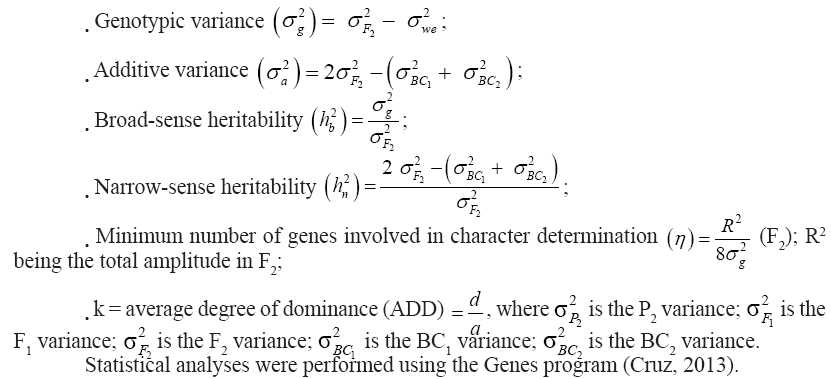
Results and Discussion
After assessing inoculations with X. euvesicatoria, no HR was evident for all evaluated generations. Therefore, the interaction between plant and pathogen was compatible (Bergamin Filho and Amorim, 2002). From the scoring scale, the results showed that all P1 parent plants reached an incidence of 100%, i.e., all plants were susceptible (Figure 4A). These findings corroborate those reported in preceding studies such as Moreira et al. (2013a, 2015). Moreover, P2 had its resistance confirmed (Figure 4B), as already observed by Moreira et al. (2010, 2013b) and Pimenta et al. (2016). It is noteworthy emphasizing that highly contrasting parents are essential for the evaluated characteristic. In F1 generation, 100% of the plants developed susceptibility symptoms (Figure 4C), indicating a recessive genetic control for resistance.
F2 plants showed various symptoms (Figure 4D, E, F, G, and H), characterizing a large genetic variability of this generation. Aggressiveness was noticed by a fast appearance of symptoms in susceptible plants. In BC1 and BC2, susceptibility rates were between 92.5 and 82.5%, respectively. These values indicate that a single gene might possibly control such a characteristic.
Resistance quantitative analysis was made considering the AUDPC values. P2 mean (13.80) was lower than that of P1 (26.30). It confirms this genotype resistance since the smaller the affected area, the more resistant is a genotype (Table 1). Riva-Souza et al. (2009) and Moreira et al. (2015), evaluating plants at the same time, also confirmed resistance by lower means for the same resistant parent, which were 15.67 and 15.5, respectively. Likewise, Demosthenes and Bentes (2011), evaluating resistance to bacterial wilt in of Capsicum accessions, reported plants with lower AUDPC, being classified as resistant. F1, F2, BC1, and BC2 showed AUDPC averages close to their susceptible parent (Table 1).
| Generation | Number of evaluated plants | AUDPC | |
|---|---|---|---|
| Mean (m) | Variance (σ2) | ||
| P1 | 20 | 26.30 | 9.62 |
| P2 | 20 | 13.80 | 0.85 |
| F1 | 20 | 22.98 | 19.38 |
| F2 | 200 | 21.17 | 30.46 |
| BC1 | 40 | 24.53 | 21.82 |
| BC2 | 40 | 20.43 | 23.94 |
Table 1: Number of evaluated plants, averages and variances for the area under disease progression curve (AUDPC) of different generations from the crossing of UENF 2285 x UENF 1381 accessions of Capsicum annuum var. annuum.
F2 reached a higher phenotypic variance regarding AUDPC (30.46), as already expected (Table 1). This generation receives greater influence from both genetic (σ2g) and environmental (σ2e) factors because of a high allelic combination between individuals. Therefore, σ2g and σ2e were 25.22 and 5.23, respectively (Table 2). In this case, genetic variation (sum of additive + dominance), which is important to estimate inheritability, registered a higher value than the environmental one. Moreira et al. (2015), studying C. annuum recombinant inbred lines, also observed σ2g values higher than the σ2e ones (172.4 and 17.1, respectively), showing that genotype has more influence than the environment on the expression of AUDPC.
| Genetic parameters | Estimates of bacterial spot resistance |
|---|---|
| Phenotypic variance (σ2f) | 30.46 |
| Environmental variance (σ2e) | 5.23 |
| Genotypic variance (σ2g) | 25.22 |
| Additive variance (σ2a) | 15.15 |
| Variance of dominance (σ2d) | 10.07 |
| Broad-sense heritability (h2b%) | 82.81 |
| Narrow-sense heritability (h2n%) | 49.74 |
| Average degree of dominance (ADD) | 0.47 |
| Maximum value in F2 | 31.00 |
| Minimum value in F2 | 11.50 |
| Minimum number of genes (η) | 4.56 |
| Genotypic determination (R2) | 86.36 |
Table 2: Estimates of genetic parameters based on AUDPC values to evaluate resistance to bacterial spot (Xanthomonas euvesicatoria) in genotypes of different generations from crosses between Capsicum annuum var. annuum accessions (UENF 2285 and UENF 1381).
There was transgressive segregation in F2 generation, with a maximum value of 33.0 and a minimum of 9.5 (Table 2). These values are outside the upper and lower patterns of the parents, evidencing that more than one gene controls the resistance to X. euvesicatoria.
The variances of the assessed genetic parameters suggested that more than four genes control this bacterial resistance inheritance (Table 2). The greater the number of genes involved in controlling an specific character, the higher the number of genotypic combinations within a population and generations are required to achieve full homozygosity (Baldissera et al., 2014).
Jones et al. (2002) and Riva et al. (2004) assessed different Capsicum genotypes and identified two (bs5 and bs6) and three recessive genes, respectively. Lobo et al. (2005), evaluating the same disease in tomato accessions, noted that the number of genes ranged from four to eight, indicating a polygenic inheritance, based on the used genotypes and crossing combinations among them. As the number of genes increased, there is a rise in the number of phenotypes, reducing the differences among them. F2 segregation tends to a continuous distribution. It also reduces each allele contribution to a given character.
Broad-sense heritability (h2b) was 82.81%, and the narrow-sense one (h2n) was 49.74% (Table 2). It means that nearly 83% of the total variance in F2 is under genetic control and about 50% comes from additive genetic effects. Riva et al. (2004) evaluated inheritance of bacterial spot resistance in Capsicum from crossing between a susceptible parent and a resistant one (UENF 1381). These authors verified broad- and narrow-sense heritability with values close to those found here, being of 82, 54 and 50.17%, respectively. The high heritability estimates and the additive gene effect relevance are favorable to the improvement of traits controlled by several genes.
There is a trend of h2b being higher than h2n, since the first reflects both additive and non-additive variances, while the second considers only the additive component. Studying the inheritance of resistance to tomato blight, Abreu et al. (2008) observed values of 54.86 and 9.06% for h2b and h2n, respectively; hence, heritability values were lower. It highlights a most intense environmental influence on tomato blight than it was on the bacterial spot.
Genotypic variance has to be studied for breeding programs to be successful. Knowing the genotype variations allows us to understand the importance of certain genetic factors for a given population (Amaral et al., 1996). By means of this variable, three major components can be measured (Fisher, 1918): additive variance (mean effects of genes), dominance (interaction between alleles within the same locus), and epistatic (interaction between alleles at different loci).
Additive variance (15.15) was superior to dominance variance (10.07) (Table 2). These outcomes indicate a high covariance between progeny and its respective parents, what implies in possible selection gains. Additive variance is a key tool for breeders since it enables the selection of a most efficient breeding method for fixation of a characteristic of interest (Cruz et al., 2014). Riva-Souza et al. (2007), studying the same pathosystem, pinpointed dominance deviations (1.11) superior to additive effects (0.32), in this case, making the selection difficult. Although studying the same pathosystem, the estimates obtained for each genetic parameter for the same trait is unique because depends on the genetic of the parents.
ADD was 0.47, expressing that genotypic value of homozygous was lower than that of heterozygous, consisting of a partial dominance with prospective epistatic effect. Riva et al. (2004), studying the same pathosystem, identified an ADD of 1.13. Similarly, Bento et al. (2013), evaluating the resistance of C. baccatum to PepYMV, verified an ADD of 1.12, indicating an overdominant action. Juhász et al. (2008) observed a value of 1.0 in a tomato-PepYMV pathosystem, indicating complete dominance. ADD is estimated by spotting a heterozygous position in relation to its contrasting homozygous parents; thus, ADD values may be different, even in a similar pathosystem.
By the coefficient of determination (R2), gene effects on resistance to the bacterial spot were confirmed as additives. This effect is explained by 86.36% of the total variation (Table 2). Moreira et al. (2015), evaluating the same disease in C. annuum recombinant inbred lines, found R2 higher than 90%, what was associated with the use of a late generation (F7). With advanced generations, R2 is associated to h2n because genotypes become pure lines; therefore, variability is attributed to an additive action, which undergoes duplication (2σ2a). High R2 values result in higher accuracy in selection of superior lineages for resistance to bacterial spot, maximizing thus genetic gains (Ribeiro et al., 2009).
The mean generation analysis showed that mean (m), additivity (a), and dominance (d) were significant which means that additive-dominant model could explain genetic effects involved in the inheritance of resistance. Mean values had the greatest estimative value (19.90) (Table 3). The t-test showed significance only for the average (m). Besides, the additive effect was higher (6.06) compared to the dominance effect (3.31), similarly to the results observed by Costa et al. (2002) in the same pathosystem. Since reduced model (additive-dominant) was sufficient to explain AUDPC genetic effects, complete model that includes epistasis data (aa, ad, and dd) was not discussed here.
| Parameter | AUDPC | ||
|---|---|---|---|
| Estimative | Variance | t | |
| m | 19.90 | 0.11 | 59.66ns |
| a | 6.06 | 0.11 | 18.22ns |
| d | 3.31 | 0.61 | 4.24* |
m - mean, a - additivity and d - dominance. nsNon-significant, *significant at P ≤ 0.05 by the t-test.
Table 3: Estimation of genetic effects for resistance to bacterial spot, in a partial model (m, a, d), for generations P1, P2, F1, F2, BC1, and BC2 from the crossing between UENF 2285 and UENF 1381 accessions of Capsicum annuum var. annuum.
In Solanaceae, a greater magnitude of the additive over the dominant effect in resistance to bacterial blight was observed in tomato segregating population (Lobo et al., 2005) and also in C. annuum (Riva et al., 2004), similar to that found on this study. This result is quite important in a breeding program since additive effects are the ones that result in fixation of the traits along with advanced generations.
Conclusion
Five genes control resistance to bacterial spot in populations derived from the crossing between accessions UENF 2285 and UENF 1381. Genetic control of bacterial spot has a quantitative aspect, with higher additive effect. Therefore, it is recommended to use breeding methods that allow selection of most advanced generations when the traits are already fixed, thus reducing the environmental effects.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgment
The authors want to thank FAPERJ for financial support and for the Ph.D. fellowship granted to L.R.A. Silva.
About the Authors
Corresponding Author
R. Rodrigues
Laboratório de Melhoramento Genético Vegetal, Universidade Estadual do Norte Fluminense Darcy Ribeiro, Campos dos Goytacazes, RJ, Brasil
- Email:
- rosana@uenf.br
References
- Abreu FB, Silva DJH, Cruz CD and Mizubuti ESG (2008). Inheritance of resistance to Phytophthora infestans (Peronosporales, Pythiaceae) in a new source of resistance in tomato (Solanum sp. (formerly Lycopersicon sp.), Solanales, Solanaceae). Genet. Mol. Biol. 31: 493-497. http://dx.doi.org/10.1590/S1415-47572008000300016
- Aguiar LA, Kimura O, Castilho AMC, Castilho KSC, et al. (2000). Resistência ao cobre em isolados nacionais de Xanthomonas campestris pv. vesicatoria de pimentão e tomateiro. Agronomia 34: 78-82.
- Amaral AL, Carvalho FIF, Federizzi LC, Mittelmann A, et al. (1996). Estimativa de herdabilidade para os caracteres adaptativos ciclo e estatura de planta em aveia. Cienc. Rural 26: 33-37. http://dx.doi.org/10.1590/S0103-84781996000100007
- Associação Brasileira de Saúde Coletiva Dossiê - ABRASCO (2015). Um Alerta Sobre os Impactos dos Agrotóxicos na Saúde. Parte 1 - Agrotóxicos, Segurança Alimentar e Nutricional e Saúde (Carneiro FF, Augusto LGS, Rigotto RM, Friedrich K, et al., eds.). Rio de Janeiro.
- Baldissera JNC, Valentini G, Coan MMD, Guidolin AF, et al. (2014). Fatores genéticos relacionados com a herança em populações de plantas autógamas. Rev. Cienc. Agrovet. 13: 181-189.
- Bento CS, Rodrigues R, Gonçalves LSA, Oliveira HS, et al. (2013). Inheritance of resistance to Pepper yellow mosaic virus in Capsicum baccatum var. pendulum. Genet. Mol. Res. 12: 1074-1082. http://dx.doi.org/10.4238/2013.April.10.3
- Bergamin Filho A and Amorim L (2002). Doenças com período de incubação variável em função da fenologia do hospedeiro. Fitopatol. Bras. 27: 561-565. http://dx.doi.org/10.1590/S0100-41582002000600001
- Castellani A (1939). Viability of some pathogenic fungi in distilled water. J. Trop. Med. Hyg. 42: 225.
- Cook and Stall (1982). Distribution of races of Xanthomonas vesicatoria pathogenic on pepper. Plant Dis. 66: 388-389. http://dx.doi.org/10.1094/PD-66-388
- Costa RA, Rodrigues R and Sudré CP (2002). Resistência genética à mancha-bacteriana em genótipos de pimentão. Hortic. Bras. 20: 86-89.http://dx.doi.org/10.1590/S0102-05362002000100016
- Cruz CD (2013). GENES - A Software Package for Analysis in Experimental Statistics and Quantitative Genetics. Acta Sci. Agron. 35: 271-276.http://dx.doi.org/10.4025/actasciagron.v35i3.21251
- Cruz CD, Regazzi AJ and Carneiro PCS (2014). Modelos Biométricos Aplicados ao Melhoramento Genético. 3° ed., v2. Viçosa, MG. Ed. UFV. 14.
- Demosthenes LCR and Bentes JLS (2011). Fontes de resistência à murcha bacteriana em germoplasma de Capsicum spp. do estado do Amazonas. Acta Amazon. 41: 435-438. http://dx.doi.org/10.1590/S0044-59672011000300016
- Filgueira FAR (2012). Novo Manual de Olericultura: Agrotecnologia Moderna na Produção e Comercialização de Hortaliças. Editora UFV, 3° ed.
- Fisher DS (1918). The correlation between relatives on the supposition of Mendelian inheritance. Trans. R. Soc. Edinb. 52: 399-433. http://dx.doi.org/10.1017/S0080456800012163
- Jones JB, Stall RE and Bouzar H (1998). Diversity among xanthomonads pathogenic on pepper and tomato. Annu. Rev. Phytopathol. 36: 41-58.http://dx.doi.org/10.1146/annurev.phyto.36.1.41
- Jones JB, Minsavage GV, Roberts PD, Johnson RR, et al. (2002). A non-hypersensitive resistance in pepper to the bacterial spot pathogen is associated with two recessive genes. Phytopathology 92: 273-277. http://dx.doi.org/10.1094/ PHYTO.2002.92.3.273
- Juhász ACP, Silva DJH, Zerbini Júnior FM, Carneiro PCS, et al. (2008). Base genética da resistência de um acesso de tomate silvestre ao mosaico-amarelo do pimentão. Pesqui. Agropecu. Bras. 43: 713-720. http://dx.doi.org/10.1590/ S0100-204X2008000600007
- Kurozawa C and Pavan MA (2005). Doenças do Tomateiro (Lycopersicon esculentum). In: Manual de Fitopatologia: Doenças das Plantas Cultivadas. 4th edn. (Kimati H, Amorim L, Rezende JAM, Bergamin Filho A, et al., eds.). São Paulo, Agronômica Ceres 2: 607-626.
- Lobo VLS, Giordano LB and Lopes CA (2005). Herança da Resistência à Mancha-Bacteriana em Tomateiro. Fitopatol. Bras. 30: 343-349.http://dx.doi.org/10.1590/S0100-41582005000400002
- Moreira SO, Rodrigues R, Araújo ML, Riva-Souza EM, et al. (2010). Desempenho agronômico de linhas endogâmicas ecombinadas de Capsicum annuum L. em sistema orgânico sob cultivo protegido. Cienc. Agrotec. 34: 886-891. http://dx.doi.org/10.1590/S1413-70542010000400013
- Moreira SO, Gonçalves LSA, Rodrigues R, Sudré CP, et al. (2013a). Correlações e análise de trilha sob multicolinearidade em linhas recombinadas de pimenta (Capsicum annuum L.). Agraria 8: 15-20. http://dx.doi.org/10.5039/agraria.v8i1a1726
- Moreira SO, Rodrigues R, Oliveira HS, Medeiros AM, et al. (2013b). Phenotypic and genotypic variation among Capsicum annuum recombinant inbred lines resistant to bacterial spot. Genet. Mol. Res. 12:232-1242.http://dx.doi.org/10.4238/2013.April.17.2
- Moreira SO, Rodrigues R, Sudré CP and Riva-Souza E (2015). Resistência à mancha-bacteriana e características agronômicas de linhas recombinadas de Capsicum annuum L. Agraria 10: 198-204. http://dx.doi.org/10.5039/ agraria.v10i2a4075
- Pedlowski MA, Canela MC, Terra MAC and Faria RMR (2012). Modes of pesticides utilization by Brazilian smallholders and their implications for human health and the environment. Crop Prot. 31: 113-118. http://dx.doi.org/10.1016/j. cropro.2011.10.002
- Pimenta S, Rodrigues R, Sudré CP, Moraes JGT, et al. (2016). Protecting vegetable cultivars in Brazil: a chili pepper case- study research. Hortic. Bras. 34: 161-167. http://dx.doi.org/10.1590/S0102-053620160000200003
- Potnis N, Minsavage G, Smith JK, Hurlbert JC, et al. (2012). Avirulence proteins AvrBs7 from Xanthomonas gardneri and AvrBs1.1 from Xanthomonas euvesicatoria contribute to a novel gene-for-gene interaction in pepper. Mol. Plant Microbe Interact. 25: 307-320.http://dx.doi.org/10.1094/MPMI-08-11-0205
- Potnis N, Timilsina S, Strayer A, Shantharaj D, Barak JD, Paret ML, Vallad GE, Jones JB (2015). Bacterial spot of tomato and pepper: diverse Xanthomonas species with a wide variety of virulence factors posing a worldwide challenge. Mol. Plant. Pathol. 16: 907-920.
- Rêgo ER, Finger FL and Rêgo MM (2011). Produção Genética e Melhoramento de Pimentas (Capsicum spp.) Areia-PB, 54.
- Ribeiro EH, Pereira MG, Coelho KS and Freitas Júnior SP (2009). Estimativas de parâmetros genéticos e seleção de linhagens endogâmicas recombinantes de feijoeiro comum (Phaseolus vulgaris L.). Rev. Ceres 56: 580-590.
- Riva EM, Rodrigues R, Pereira MG, Sudré CP, et al. (2004). Inheritance of bacterial spot disease in Capsicum annuum L. Crop Breed. Appl. Biotechnol. 4: 490-494.http://dx.doi.org/10.12702/1984-7033.v04n04a18
- Riva-Souza EM, Rodrigues R, Sudré CP, Pereira MG, et al. (2007). Obtaining pepper F2:3 lines with resistance to the bacterial spot using the pedigree method. Hortic. Bras. 25: 567-571. http://dx.doi.org/10.1590/S0102-05362007000400014
- Riva-Souza EM, Rodrigues R, Sudré CP, Gonzaga MP, et al. (2009). Genetic parameters and selection for resistance to bacterial spot in recombinant F6 lines of Capsicum annuum. Crop Breed. Appl. Biotechnol. 9: 108-115. http://dx.doi. org/10.12702/1984-7033.v09n02a02
- Rodrigues Neto J, Malavolta Jr VA and Victor O (1986). Meio simples para o isolamento e cultivo de Xanthomonas campestris pv. citri tipo B. Summa Phytopathol. 12: 16.
- Shaner G and Finney RE (1977). The effect of nitrogen fertilization on the expression of slow-mildewing resis/ance in knox wheat. Phytopathology 70: 1183-1186. http://dx.doi.org/10.1094/Phyto-70-1183
- Stall RE, Jones JB and Minsavage GV (2009). Durability of resistance in tomato and pepper to xanthomonads causing bacterial spot. Annu. Rev. Phytopathol. 47: 265-284. http://dx.doi.org/10.1146/annurev-phyto-080508-081752
- Sudré CP, Gonçalves LSA, Rodrigues R, do Amaral Júnior AT, et al. (2010). Genetic variability in domesticated Capsicum spp as assessed by morphological and agronomic data in mixed statistical analysis. Genet. Mol. Res. 9: 283-294. http://dx.doi.org/10.4238/vol9-1gmr698
- Vallejos CE, Jones V, Stall RE, Jones JB, et al. (2010). Characterization of two recessive genes controlling resistance to all races of bacterial spot in peppers. Theor. Appl. Genet. 121: 37-46. http://dx.doi.org/10.1007/s00122-010-1289-6
Keywords:
Download:
Full PDF- Share This



