Large-scale development, characterization, and cross-amplification of EST–SSR markers in Chinese chive
Received: October 14, 2017
Accepted: November 08, 2017
Published: January 05, 2018
Genet.Mol.Res. 17(1): gmr16039861
DOI: 10.4238/gmr16039861
Abstract
Chinese chive (Allium tuberosum Rottler ex Spr.) is an important vegetable crop. However, genetic and breeding studies of the species have been restricted by the lack of simple sequence repeat markers (SSRs). In the present study, a total of 2553 Chinese chive SSRs were developed from the species’ transcriptome, with 626, 643, and 536 of the makers located in coding sequences, 5' untranslated regions and 3' untranslated region, respectively. The annotation of SSR-containing expressed sequence tags revealed that the transcripts were enriched for several Gene Ontology (GO) categories, including ‘protein binding’, ‘regulation of transcription’, and ‘integral to membrane’. Among the 2,553 SSRs, di- and tri-nucleotide repeat motifs were the most abundant (52.3 and 45.6%, respectively), and AC/GT and GAA/TTC were the most frequent di- and tri-nucleotide motifs, respectively. PCR amplification, using 100 SSR primer pairs, revealed that 94% of the markers were of good quality and that 83-88 of the makers could be amplified in six other Allium species. This suggests that the markers had high cross-species transferability. The substantial number of SSRs developed will provide a valuable resource for future genetic and breeding studies in Chinese chive
Introduction
Chinese chive (A. tuberosum Rottler ex Spr.) is a tetraploid (2n = 4X = 32) perennial that belongs to the Liliaceae, and the species contain an abundance of organic sulfur compounds, which are responsible for plant’s characteristic flavours and beneficial properties (Mau et al., 2001). As an economically important crop, Chinese chive is consumed, both as a vegetable and spice, and is also widely used in traditional medicine for treating a variety of diseases (Imahori et al., 2007). Because of the species’ nutritional and medicinal properties, it is widely on a global scale, especially in China, Korea, and Japan.
Nevertheless, despite the economic importance of Chinese chive, a poor understanding of the crop’s genetics has hindered the improvement of crop traits. To date, no genetic maps and quantitative trait loci (QTLs) have been reported. Several markers without not specific loci, such as inter simple sequence repeat (ISSR) and random amplified polymorphic DNA (RAPD) makers, have been used in the species (Pan et al., 2005), but both ISSR and RAPD markers share a variety of common shortcomings, such as poor repeatability and the effects of dominance. In contrast, simple sequence repeat (SSR) markers are recognized for several advantages, including their specific locations in the genome, high reproducibility, and multiallelic and codominant properties, as well as for the simplicity of analysis (Liu et al., 2015). As a result, SSRs are commonly used in a broad range of crop research areas, including genetic diversity analysis , genetic mapping, and crop-breeding (Liu et al., 2009, 2010a, 2011a; Mao et al., 2011; Li et al., 2012).
The identification of SSRs typically involves large-scale sequencing of whole genomes, SSR-enriched genome regions or expressed sequence tag (EST) libraries. Given recent developments in sequencing technology, large numbers of EST-derived SSR markers have been developed de novo assembly of species transcriptomes (Ding et al., 2015), and these makers have been used extensively in the genetic and breeding studies of crops (Zhu et al., 2016). however, even though the Chinese chive transcriptome has been de novo assembled (Zhu et al., 2017a) and various SSRs have been developed (Zhou et al., 2015), the number of available SSR marker is insufficient. Therefore, to improve the resources available for genetic and breeding studies of Chinese chive, the aim of the present study was to complete the large-scale development of transcriptome-based SSR markers for Chinese chive.
Materials and Methods
Plant materials, and nucleic acid extraction
Chinese chive (A. tuberosum) and six congeners (leek, Chinese jiaotou, shallot, onion, garlic, and Welsh onion). were planted in an experimental field that belongs to the Institute of Bast Fiber Crops, Chinese Academy of Agricultural Sciences, Changsha, China, on September 3, 2014. Fresh leaves of the plants were collected and subject to DNA extraction, using a Plant Genomic DNA Kit (TIANGEN, China).
Identification of SSR loci and development of markers
The Chinese chive transcriptome sequences (accession number of GFAN00000000) were downloaded from the GenBank database. SSR motifs were identified from the Chinese Chive ESTs using MISA 1.0, with default parameters: a minimum of six repeats for dinucleotide motifs, and five repeats for trinucleotide, tetranucleotide, and pentanucleotide motifs. Primer 3.0 was used to design primer pairs that flanked the putative SSRs, with the following parameters: length, 17–23 bp; GC content, 40–60%; and estimated amplicon size, 100–300 bp. To determine the location of SSR in EST, the coding sequence (CDS) of each transcript was predicted by BLAST searching against NCBI non-redundant protein sequence and Swiss Prot protein databases, as well as by EST Scan. WEGO was used to subject the SSR-containing sequences to GO functional classification, and enrichment analysis was performed using GOseq and the Wallenius noncentral hypergeometric distribution. Q values were used to determine the P-value threshold in multiple tests and analysis, and GO categories with Q-values < 0.05 were considered significantly enriched.
Amplification of SSR-containing loci
To assess the quality and cross-species transferability of the SSR flanking primer pairs, we attempted to amplify one hundred SSR markers from all seven Allium species. PCR amplification was performed in 20-μl reactions that each included 2 μl genomic DNA (30 ng/μl), 1.6 μl dNTP mix (2.5 mM), 1.2 μl of the specific primer pair(10 pmol/μl), 2.0 μl 10× rTaq PCR buffer (Laifeng, shanghai, China), and 0.4 μl rTaq polymerase (5 unit/μl; Laifeng), and the PCR conditions were as follows: 94 °C (3 min); 34 cycles of 94 °C (30 s), 56 °C (45 s), and 72 °C (45 s); and a final extension step at 72 °C (5 min). The SSR assay was performed as described by Wu and Tanksley (1993); briefly, SSRs that could also be amplified at least one of the other six Allium species was transferable.
Results
SSR markers
SSR mining identified a total of 4235 SSR loci from the 148,715 EST sequences (83.78 Mb.) Therefore, the frequency of occurrence for the EST-SSRs was one SSR per 19.8 kb of EST sequence. Of the 4235 loci, 1682 were located at the end of an EST sequence, which made it difficult to design the flanking primer pairs, and as a result, those loci were not developed into markers. For the remaining 2553 loci primer pairs (CCM0001 to CCM2533, respectively; Table S1) were designed.
Maker characterization
Of the 2553 SSR markers, dinucleotide repeat motifs were the most abundant type (1335, 52.3%), followed by trinucleotide repeat motifs (1164, 45.6%); However, only 49 tetranucleotide, 3 pentanucleotide, and 2 hexanucleotide SSR markers were identified (Table 1). For the makers with di- to hexanucleotide motifs, SSR length (number of repeat units) was determined (Table 1). Most of the SSRs (1,885 SSRs, 73.8%) ranged from 12 to 15 bp in length, whereas most of the remaining markers (552 SSRs, 20.4%) ranged from between 16 and 18 bp in length.
| Number of repeat units | Dinucleotide | Trinucleotide | Tetranucleotide | Pentanucleotide | Hexanucleotide |
|---|---|---|---|---|---|
| 5 | 818 | 40 | 2 | 1 | |
| 6 | 707 | 263 | 8 | 1 | 1 |
| 7 | 330 | 66 | 1 | ||
| 8 | 182 | 15 | |||
| 9 | 77 | ||||
| 10 | 30 | 1 | |||
| 11 | 9 | ||||
| 12 | 1 | ||||
| Total | 1335 | 1164 | 49 | 3 | 2 |
Table 1. Length distribution of EST-SSRs based on the number of repeat units.
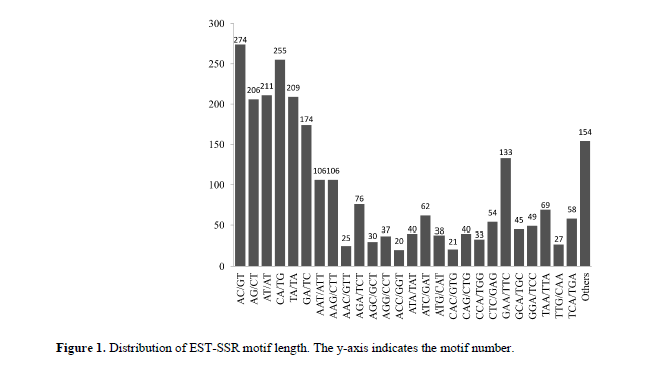
Among the 63 motif sequence types that were identified. Among these, eight, 30, 20, three, and two were di, tri-, tetra-, penta-, hexanucleotide repeat motifs, respectively. Of the eight dinucleotide repeat motifs, the AC/GT motif was the most common (274, 10.74%), followed by the CA/TG (255, 9.99%), AT/AT (211, 8.27%), TA/TA (209, 8.19%), AG/CT (206, 8.07%), and GA/TC (174, 6.82%) motifs (Figure 1). Among the 30 trinucleotide repeats motifs, the GAA/TTC, AAT/ATT, and AAG/CTT motifs were three most abundant (133, 106, and 106 SSRs, respectively), whereas the frequencies of the other motifs were all < 100 (Figure 1).
Annotation of SSR-containing ESTs
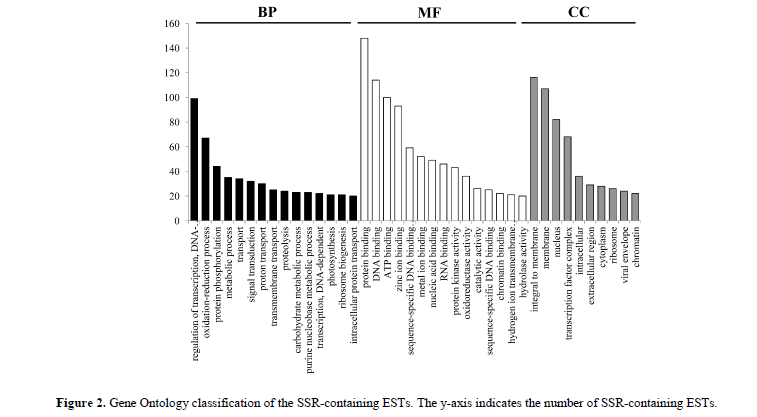
The 2553 SSRs were distributed among 2476 EST sequences, 626, 643, and 536 SSRs located in the CDS regions, 5' untranslated regions (5' UTRs), and 3' untranslated regions (3' UTRs), respectively (Table S1). Because it is impossible to determine the CDS regions of some ESTs, the location of 748 SSRs on their respective ESTs was uncertain. Of the 2476 SSR-containing ESTs, 1326 had already been annotated by Zhu et al. 2017a, (Table S2), and 179 those had been annotated as transcription factors. GO functional classification also matched the 2476 SSR-containing sequences to known proteins in the GO database, with a total of 4332 functional terms. Of those terms, 1642, 1764, and 926 were assigned to the ‘biological process’, ‘molecular function’, and ‘cellular component ontologies’, respectively (Figure 2). Interestingly, there was a significant enrichment observed in several categories namely ‘protein binding’, ‘regulation of transcription’, and integral to membrane’ (Q < 0.05).]
Only the functional categories with more than 20 terms are shown, and the black, white, and gray histogram bars represented the three most common categories: biological process (BP), molecular function (MF), and cellular component (CC).
SSR marker quality and transferability
Of the 100 tested primer pairs, 94 (94%) facilitated successful amplification, which suggested that the novel SSR markers were of good quality, and 84, 83, 84, 88, 86 and 86 of the primer pairs were also able to amplify PCR products from garlic, leek, Welsh onion, Chinese jiaotou, shallot, and onion, respectively (Table S3). In fact, 73 of the 100 Chinese chive markers were transferable to all six other Allium species, and 92 of the markers could produce DNA fragments in at least one of the six other Allium species.
Discussion
Development of 2,553 SSRs in Chinese chive
In crop research, SSR markers are an important tool for genetic and breeding studies, such as QTL identification (Zhang et al., 2009; Liu et al., 2010b, 2011b) and genetic diversity analysis (Liu et al., 2013; Zhu et al., 2017b). However, few SSR markers can have been reported for Chinese chive, mostly owing to the crop’s giant genome. Unlike genomic sequencing, transcriptome analysis is rapid, inexpensive, and unaffected by genome size or complexity and, as such, has been widely used as a primary tool for gene discovery (Zheng et al., 2015), investigating crop domestication patterns (Zeng et al., 2016), and gene expression profiling (Yu et al., 2015; Mei et al., 2016;), especially for the large-scale development of SSR markers (Ding et al., 2015). In the present study, we developed 2533 SSRs using the transcriptome sequence of Chinese chive, and 94% of the makers could successfully amplify their target sequences. The substantial number of SSRs developed here will accelerate both genetic and breeding studies of Chinese chive.
It is noted that SSR variations in the expressed sequences might have important effects on gene function. For example, expansions or contractions of SSRs in CDS regions are likely to cause frame shifts or indels in protein-encoding sequences, thereby disrupting the function of the encoded protein, and changes in the length of SSRs in 5' and 3' UTRs could affect gene expression and transcription slippage, respectively. In the present study, 626, 643, and 536 of the 2552 SSRs were in CDS regions, 5' UTRs, and 3' UTR, respectively. In addition, most of the 2476 ESTs were annotated to have important functions. For example, at least 179 of the SSR-containing sequences encoded transcription factors. and, thus, the total pool of ESTs was significantly enriched in several important categories, including ‘transcription regulation’. Because transcription factors have central roles in the regulation of plant growth, development, and stress response (Zheng et al., 2016a, b), these SSR markers will be useful for selecting and pyramiding agriculturally valuable alleles during molecular marker-assisted Chinese chive breeding programs.
Comparison of SSR features in Chinese chive and other species
In the present study, the mean distance between the identified EST-SSRs (19.8 kb) suggests that EST-SSRs are less prevalent in Chinese chive than in many other crops (Ding et al., 2015) but more prevalent than in its congener, garlic (Liu et al., 2015). In addition, the di- and tri-nucleotide repeat motifs were the most abundant, which is like observations in other plant species (Guo et al., 2014; Ding et al., 2015). However, it is important to note that the mean distance between SSRs, and motif abundances, are both dependent on SSR search criteria and the sizes of databases being searched (Varshney et al., 2005).
In Chinese chive, the AC/GT and GAA/TTC motif sequences were the most frequent di- and trinucleotide motifs, respectively, which is like the motif sequences reported for garlic (Liu et al., 2015). Previous studies have reported that GC-rich SSR motifs are more prevalent in ESTs from monocots than those from dicots, and that the abundance of CCG/CGG motifs is a common characteristic of monocot genomes (Peng and Lapitan, 2005). However, among the 2553 Chinese chive SSR makers identified in the present study, only 42 were GC-composite motifs, whereas 661 were AT-composite motif, which suggests that AT-rich motifs are more prevalent than GC-rich motifs in the EST-SSRs of this species. A similar phenomenon was observed in garlic (Liu et al., 2015). Therefore, even though Allium is a monocot genus, GC-rich SSR motifs are less prevalent in the ESTs of Allium species than in other monocot species.
Cross-species transferability of SSRs
Of 100 EST–SSRs developed in Chinese chive, 92 were transferable to at least one of the other six other Allium species investigated, and 73 were transferable to all six. This high transferability is consistent with previous reports of SSR transferability in plants (Guo et al., 2014), and the high transferability of SSRs among Allium species has also been reported for garlic SSRs (Liu et al., 2015). 87.5% garlic SSRs were transferable to Chinese chive (Liu et al., 2015), whereas, in the current study, we found that 84% of the Chinese chive SSRs were transferable to garlic. The transferability of such markers is dependent on the ability of the SSR-flanking primer pairs to bind the appropriate regions of other species. Therefore, the high transferability of Chinese chive SSRs to other Allium species suggests that the genomic sequences of these Allium species are highly conserved, and that the Allium species investigated in this study could be closely related. Furthermore, it is also likely that the SSR makers developed in the present study could also be used for genetic and breeding studies in other Allium species.
Acknowledgments
This work was supported by grants from The Agricultural Science and Technology Innovation Program of China (CAAS-IBFC-ASTIP).
About the Authors
Corresponding Author
Fu Li
Institute of Bast Fiber Crops, Chinese Academy of Agricultural Sciences, Changsha, China
- Email:
- lewterme@webmail.hzau.edu.cn
References
- Ding X, Jia Q, Luo X, Zhang L, et al. (2015). Development and characterization of expressed sequence tag-derived simple sequence repeat markers in tropical forage legume Stylosanthes guianensis (Aubl.) Sw. Mol. Breeding. 35:202. https://doi.org/10.1007/s11032-015-0370-x
- Guo R, Mao Y, Cai J, Wang J, et al. (2014). Characterization and cross-species transferability of EST–SSR markers developed from the transcriptome of Dysosma versipellis (Berberidaceae) and their application to population genetic studies. Mol. Breeding. 34:1733-1746. https://doi.org/10.1007/s11032-014-0134-z
- Imahori Y, Suzuki Y, Kawagishi M, Ishimaru M, et al. (2007). Physiological responses and quality attributes of Chinese chive leaves exposed to CO2-enriched atmospheres. Postharvest Biol. Tec. 46:160-166. https://doi.org/10.1016/j.postharvbio.2007.04.008
- Li F, Ma C, Chen Q, Liu T, et al. (2012). Comparative mapping reveals similar linkage of functional genes to QTL of yield-related traits between Brassica napus and Oryza sativa. J. Genet. 91:163-170. https://doi.org/10.1007/s12041-012-0155-5
- Liu T, Mao D, Zhang S, Xu C, et al. (2009). Fine mapping SPP1, a QTL controlling the number of spikelets per panicle, to a BAC clone in rice (Oryza Sativa). Theor. Appl. Genet. 118:1509-1517. https://doi.org/10.1007/s00122-009-0999-0 Liu T, Shao D, Kovi M and
- Xing Y (2010a). Mapping and validation of quantitative trait loci for spikelets per panicle and 1000-grain weight in rice (Oryza sativa L.). Theor. Appl. Genet. 120:933-942. https://doi.org/10.1007/s00122-009-1222-z
- Liu T, Zhang Y, Xue W, Xu C, et al. (2010b). Comparison of quantitative trait loci for 1,000-grain weight and spikelets per panicle across three connected rice populations. Euphytica. 175: 383-394. https://doi.org/10.1007/s10681-010-0186-z
- Liu T, Zhang Y, Zhang H, Xing Y (2011a). Quantitative trait loci for the number of grains per panicle dependent on or independent of heading date in rice (Oryza Sativa L.). Breeding Sci. 61:142-150. https://doi.org/10.1270/jsbbs.61.142
- Liu T, Li L, Zhang Y, Xu C, et al. (2011b). Comparison of quantitative trait loci for rice yield, panicle length and spikelet density across three connected populations. J. Genet. 90:377-382. https://doi.org/10.1007/s12041-011-0083-9 Liu T, Liu H, Zhang H and
- Xing Y (2013). Validation and characterization of Ghd7.1, a major QTL with pleiotropic effects on spikelets per panicle, plant height, and heading date in rice (Oryza sativa L.). J. Integrat. Plant Biol. 55:917–927. https://doi.org/10.1111/jipb.12070
- Liu T, Zeng L, Zhu S, Chen X, et al. (2015). Large-scale development of expressed sequence tag-derived simple sequence repeat markers by deep transcriptome sequencing in garlic (Allium sativum L.). Mol. Breeding. 35:204. https://doi.org/10.1007/s11032-015-0399-x
- Mao D, Liu T, Xu C, Li X, et al. (2011). Epistasis and complementary gene action adequately account for the genetic bases of transgressive segregation of kilo-grain weight in rice. Euphytica. 180:261–271. https://doi.org/10.1007/s10681-011-0395-0
- Mau J, Chen C, and Hsieh P (2001). Antimicrobial effect of extracts from Chinese chive, cinnamon, and corni fructus. J. Agri. Food Chem. 49:183-188. https://doi.org/10.1021/jf000263c
- Mei S, Liu T and Wang Z (2016). Comparative transcriptome profile of the cytoplasmic male sterile and fertile floral buds of radish (Raphanus sativus L.). Int. J. Mol. Sci. 17:42. https://doi.org/10.3390/ijms17010042
- Pan M, Yang J, Cao D, Li X, et al. (2005). Genetic diversity of cultivated Chinese chive germplasm by ISSR and RAPD markers. Chinese Agricultural Bulletin. 21: 44-47. https://doi.org/10.3724/sp.j.1006.2011.00595
- Peng JH and Lapitan NL (2005). Characterization of EST-derived microsatellites in the wheat genome and development of eSSR markers. Funct. Integr. Genomics. 5:80-96. https://doi.org/10.1007/s10142-004-0128-8
- Varshney RK, Graner A and Sorrells ME (2005). Genic microsatellite markers in plants: features and applications. Trends Biotechnol. 23:48-55.https://doi.org/10.1016/j.tibtech.2004.11.005
- Wu KS, Tanksley SD (1993). Abundance, polymorphism and genetic mapping of microsatellites in rice. Mol. Gen. Genet. 241:225-235. https://doi.org/10.1007/bf00280220
- Yu Y, Zeng L, Yan Z, Liu T, et al. (2015). Identification of ramie genes in response to Pratylenchus coffeae infection challenge by digital gene expression analysis. Int. J. Mol. Sci. 16:21989–22007. https://doi.org/10.3390/ijms160921989
- Zeng L, Shen A, Chen J, Yan Z, et al. (2016). Transcriptome analysis of ramie (Boehmeria nivea L. Gaud.) in response to ramie moth (Cocytodes coerulea Guenée) infestation. BioMed Res. Int. 2016:3702789. https://doi.org/10.1155/2016/3702789
- Zhang Y, Luo L, Liu T, Xu C, et al. (2009). Four rice QTL controlling number of spikelets per panicle expressed the characteristics of single Mendelian gene in near isogenic backgrounds. Theor. Appl. Genet. 118:1035-1044. https://doi.org/10.1007/s00122-008-0960-7
- Zheng J, Yu C, Chen P, Wang Y, et al. (2015). Identification and expression analysis of glutamine synthetase genes in ramie (Boehmeria nivea L. Gaud). Open Life Sci. 10: 299–309. https://doi.org/10.1515/biol-2015-0032
- Zheng X, Zhu S, Tang S and Liu T (2016a). Identification of drought, cadmium and root-lesion nematode infection stress-responsive transcription factors in ramie. Open Life Sci. 11: 191–199. https://doi.org/10.1515/biol-2016-0025
- Zheng X, Tang S, Zhu S, Dai Q, et al. (2016b). Identification of an NAC transcription factor family by deep transcriptome sequencing in onion (Allium cepa L.). PLoS ONE.11: e0157871. https://doi.org/10.1371/journal.pone.0157871
- Zhou S, Chen L, Liu S, Wang X, et. al. (2015). De novo assembly and annotation of the Chinese chive (Allium tuberosum Rottler ex Spr.) transcriptome using the Illumina platform. PLoS ONE. 10: e0133312. https://doi.org/10.1371/journal.pone.0133312
- Zhu S, Zheng X, Dai Q, Tang S, et al. (2016). Identification of quantitative trait loci for flowering time traits in ramie (Boehmeria nivea L. Gaud). Euphytica. 210:367–374. https://doi.org/10.1007/s10681-016-1692-4
- Zhu S, Tang S, Tan Z, Yu Y, et al. (2017a). Comparative transcriptomics provide insight into the morphogenesis and evolution of fistular leaves in Allium. BMC Genomics. 18:60. https://doi.org/10.1186/s12864-016-3474-8
- Zhu S, Liu T, Dai Q, Wu D, et al. (2017b). Genetic structure and relationships of an associated population in ramie (Boehmeria nivea L. Gaud) evaluated by SSR markers. Biotechnol. Biotec. Eq. 31:36–44. https://doi.org/10.1080/13102818.2016.1253437
Keywords:
Download:
Full PDF- Share This