Long non-coding RNA-protein interaction: the preliminary step to track their biological functions
Received: December 11, 2017
Accepted: September 01, 2018
Published: November 05, 2018
Genet.Mol.Res. 17(4): http://dx.doi.org/gmr16039920
DOI: http://dx.doi.org/10.4238/gmr16039920
Abstract
RNA is not only an intermediate for the flow of genetic information between DNA and protein. The transcription of the entire genomes generates a myriad non-protein coding RNA in addition to protein coding genes. Among these transcripts, lncRNA which is the least well understood and they cannot be considered as merely transcriptional noises. Here, we review the emerging role of lncRNA interactions with binding proteins (RBP) to perform multidirectional functions in transcription and epigenetic gene regulations, control of mRNA splicing, translation and post translational modifications. In addition, lncRNAs may acts as cellular thermostat that helps in preserving of cellular homeostasis. Keywords: lncRNAs; RBPs; cellular hemostasis
Introduction
RNA binding proteins (RBPs) and formation of ribonucleoprotein complex.
RNAs ability to interact with others RNAs and or proteins, such as miRNA-mRNA and lncRNA-mRNA interactions, plays an important role in posttranscriptional gene regulations [Bartel, D.P et al., 2009, Gong, C et al., 2011]. However, the global prevalence and dynamic of RNA interaction networks and their impact on gene regulation are still largely unknown. Such mapping of lncRNA structure and interactomes in different cellular states is crucial for our understanding of lncRNA biology.
A transcript is usually considered as ncRNA, rather than protein, if it lacks any substantioal open reading frame (ORF), fails to produce a protein during invitro translation experiments or encodes short peptides from small open reading frames (smORFs) (Payre F et al., 2016). According to size of these ncRNAs, over than 200nt considered as lncRNAs. Thousands of these lncRNAs were identified by the recent advent of RNA deep sequencing and microarray data. To date, the detailed function of these transcripts remains unknown.
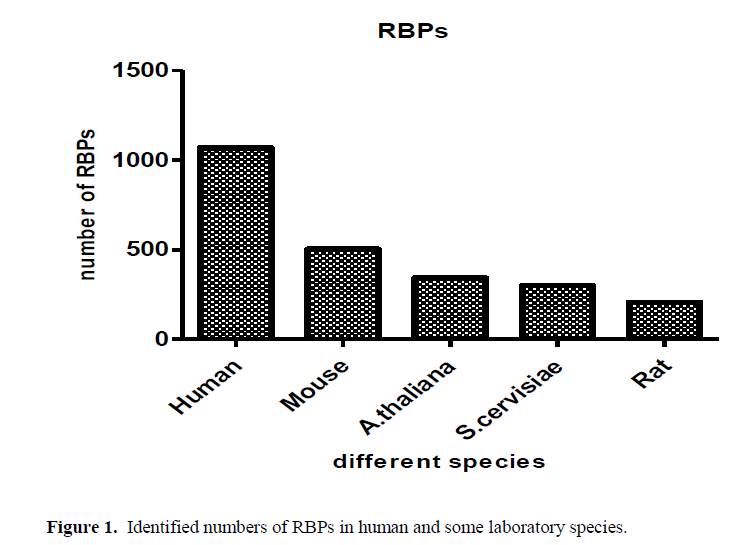
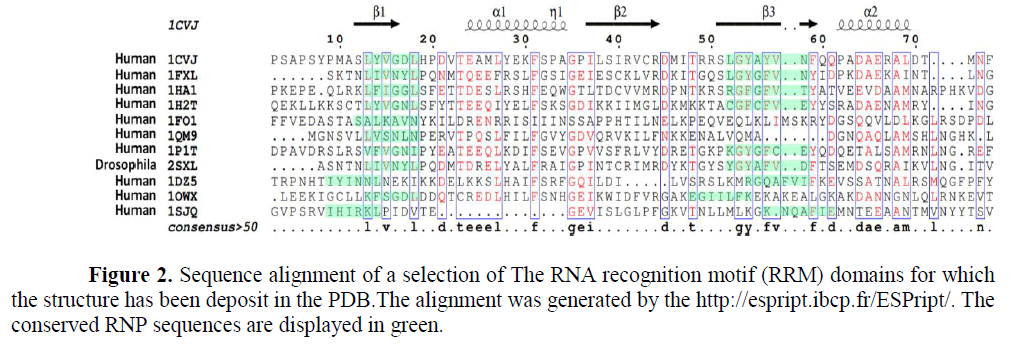
From the old saying “you can judge the person from the company he keeps”. RBP, are proteins that bind to RNA in cells and participate in forming of ribonucleoprotein complexes either these proteins are cytoplasmic or nuclear located. Thousands of RBPs were identified in human as well as different laboratory species as shown in Figure (1). Further, table (1) annotated different properties of RBPs domains. RBPs have crucial roles in various cellular processes such as: cellular function, transport and localization. They especially play a major role in posttranscriptional control of RNAs, such as: splicing, polyadenylation, mRNA stabilization, mRNA localization and translation. As it become clear not the whole sequence of protein interacts with RNA but only small Motif as shown in Figure (2) RNA recognition motifs in different species. So that, the first step to unveil the lncRNAs possible biological functions is to identify their binding partner. Our group already identified a number of lncRNAs that were deregulated in old and young mouse different organs (un-published data). Furthermore, some bioinformatics tools used to predict whether a candidate lncRNAs can interact with RBP or micRNA (Li J.H et al., 2014, Paraskevopoulou, MD et al., 2013)
| Domain √?¬†Annotation | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| PDB ID | Classification | Organism |
Family |
Domain | RNA-recognition surface | Molecular Function | RNA Sequence 5’ to 3’ |
Topology |
Reference |
| 1URN | Transcription | Homo sapiens | Canonical RBD | Splicesomal U1A protein | Surface of √?¬≤-sheet | Nucleotide Binding Nucleic Acid Binding, Snrna Binding | AAUCCAUUGCACUCCGGAUUU | Alpha-Beta Plaits | (Oubridge, C et al .,1994) |
| 1EC6 | RNA binding protein | Homo sapiens | Eukaryotic type KH-domain (KH-domain type I) | Neuro-oncological ventral antigen 2, nova- 2, KH3 | Hydrophobic cleft√?¬† created√?¬† between GXXG loop and√?¬† √?¬≤2, √?¬≤3 . Type II: same as type I, except√?¬† inconstant√?¬† loop is between√?¬† √?¬≤2 and √?¬Ī2 |
Nucleic Acid Binding RNA Binding | GAGGACCUAGAUCACCCCUC | Ribosomal Protein S8; Chain: A, domain 1 | (Lewis, H.A et al., 2000) |
| 1EKZ | Cell cycle | Drosophila melanogaster | Double-stranded RNA-binding domain (dsRBD) | Staufen, domain III | √?¬Ī1, √?¬Ī2, and loop between √?¬≤1 and√?¬≤2 | GGACAGCUGUCCCUUCGG GGACAGCUGUCC |
Double Stranded RNA Binding Domain | (Ramos A., et al., 2000) | |
| 1UN6 | RNA-binding protein | Xenopus laevis | Classic zinc finger, C2H2 | Transcription factor IIIA, TFIIIA | Primarily residues in √?¬Ī-helices | Nucleic Acid Binding Metal Ion Binding |
GCCGGCCACACCUACGGG GCCUGGUUAG UACCUGGG AAACCUGGGA AUACCAGGUGCCGGC |
Double Stranded RNA Binding Domain | (Lu, D etal., 2003) |
| 1RGO | RNA binding protein | Homo sapiens | CCCH zinc finger | Butyrate response factor 2 (Tis11D) | Aromatic side chains form hydrophobic binding pockets for bases that make direct hydrogen bonds to protein backbone |
Metal Ion Binding | UUAUUUAUU | Little regular secondary structure |
(Hudson, B.P et al., 2004) |
| 2IX1 | Hydrolase | Escherichia coli | Cold shock DNA-binding domain-like | Exoribonuclease 2, RNB | Core formed by 2√?¬≤-strands√?¬† together with√?¬† contributions from surrounding loops | 3' 5' Exoribonuclease Activity Nucleic Acid Binding RNA Binding Nuclease Activity Exonuclease Activity Ribonuclease Activity Exoribonuclease Ii Activity Hydrolase Activity |
AAAAAAAAAAAAA | Alpha-Beta Plaits | (Frazao, C., et al 2006) |
| 1SI3 | Gene regulation | Homo sapiens | PAZ domain | Eukaryotic translation initiation factor 2C 1, EIF2C1 | Hydrophobic pocket√?¬† created by OB-like √?¬≤-barrel as well as small √?¬Ī√?¬≤ motif |
CGUGACUCU | Paz domain | (Ma, J.B et al., 2005) | |
| 1C9S | RNA binding protein | Geobacillus stearothermophilus | Trp RNA-binding attenuation protein (TRAP) | Trp RNA-binding attenuation protein (TRAP) | Edges of √?¬≤-sheets√?¬† through√?¬† all of the eleven√?¬† subunits that form the entire protein structure |
RNA Binding | GAUGAGAUGAGAUGAGA UGAGAUGAGAUGAGAUGA GAUGAGAUGAGAUGAGAUGA |
Immunoglobulin-like | (Antson, AA et al.,1999) |
| 1M8Y | RNA binding protein | Homo sapiens | Pumilio repeat | Pumilio 1 | 2 repeats combine to form binding pocket for individual bases; helix √?¬Ī2 provides specificity-determining residues |
RNA Binding | AUUGUACAUA | Leucine-rich Repeat Variant | Wang, X etal., 2001) |
| 1YTU | RNA binding Protein | Archaeoglobus fulgidus | PIWI domain | Hypothetical protein AF1318 | Highly conserved pocket,√?¬† inclusive a metal ion that is bound to the exposed C-terminal carboxylate | Nucleic Acid Binding RNA Binding Metal Ion Binding |
AGACAG UGUC |
Nucleotidyltransferase; domain 5 Rossmann fold |
(Ma, J.B., et al 2005) |
Table 1: The different properties of some RBPs domains.
Proteostasis (protein homeostasis), is the process which govern protein cycle begin from biogenesis, folding, trafficking, activity, interaction, degradation and elimination. Proteostasis disruption can leads to disruption of all cellular functions and involved in many diseases, as aging and aging associated diseases as Alzheimer’s, Parkinson’s, and Huntington’s diseases (Powers ET et al., 2009, Balch WE et al., 2008) Following proteins; disruption of lncRNAs-homeostasis (lncRNAstasis) altered in cellular physiological and pathological conditions (see the comprehensive review demonstrating different lncRNAs disrupted in cellular senescence (A.R. Ghanam). So, like Proteins lncRNAs control cellular homeostasis in different levels from cell development, differentiation, migration in the embryo till programmed cell death or cellular senescence. It is clear now Proteostasis and lncstasis elicit the cell life and death decision. So that, there is overwhelming interest to explore the exact relation between Proteostasis and lncRNAstasis at both molecular level and phenotypic level.
All cells take advantage of an array of quality control mechanisms to preserve the stability and functionality of their proteomes. Proteostasis involves mechanisms for the stabilization of correctly folded proteins most prominently, the heat shock family of proteins and mechanisms for the degradation of proteins by the proteasome or the lysosome (Hartl FU et al., 2011, Koga H et al., 2011). All of these systems function in a coordinated fashion to restore the structure of misfolded polypeptides or to remove and degrade them completely, thus preventing the accumulation of damaged components and assuring the continuous renewal of intracellular proteins. Additionally, chronic expression of unfolded, misfolded, or aggregated proteins contributes to altered homeostasis and predispose to some diseases . As the first shadow of lncRNA appears in molecular biology its involvement in this quality control mechanism. So, there is overwhelming interest to explore the relation between proteostasis and lncRNAstais.
1CVJ is the poly(A)-binding protein. (Deo RC et al., 1999); 1FXL is the HuD protein. (Wang X et al., 2001) ; 1HA1 is the HNRNP. (Shamoo Y et al., 1997); 1H2T is the Nuclear CAP-Binding. (Mazza C et al., 2002) ; 1FO1 is the canonical RNP domain and an LRR domain. (Liker E et al., 2000); 1QM9 is the Polypyrimidine Tract binding protein. (Conte M.R et al., 2000); 1P1TC is the stF-64 protein. (Perez Canadillas J.M et al., 2003) ; 2SXL is The Sex-lethal (Sxl) protein. (Inoue M. et al., 1997) ; 1DZ5 is the U1A Protein. (Varani L et al., 2000); 1OWX is the human La protein. (Jacks A et al., 2003) and 1SJQ is the Polypyrimidine Tract Binding Protein Isoform 1 (PTB1). (Simpson P.J et al., 2004)
The foregoing discussion has provided a survey of the present state of knowledge regarding the role of lncRNA associated with Proteostasis in normal and disease conditions.
Nuclear lncRNAs
LncRNAs and subcellular localization of proteins, some lncRNA regulates the abundance of proteins in different subcellular compartments. By recruiting transcription factors to the nucleus, specifically to certain regions of DNA, these lncRNAs can modulate transcription. Similarly, lncRNAs can recruit certain RNA binding protein into ribonucleoprotein complexes figure (3). Although the complete mechanism through which lncRNA affect protein trafficking and their impact on cellular homeostasis are largely unknown. Depending in their site of action, they may be divided into cis- and trans-acting transcripts. The cis-acting lncRNAs modulate expression of neighboring genes located in close proximity to lncRNA genic region on the same chromosome. In contrast, trans-acting lncRNAs regulates transcription of genes located on other chromosomes (Rinn, J.L et al., 2012, Guttman, M et al., 2012).
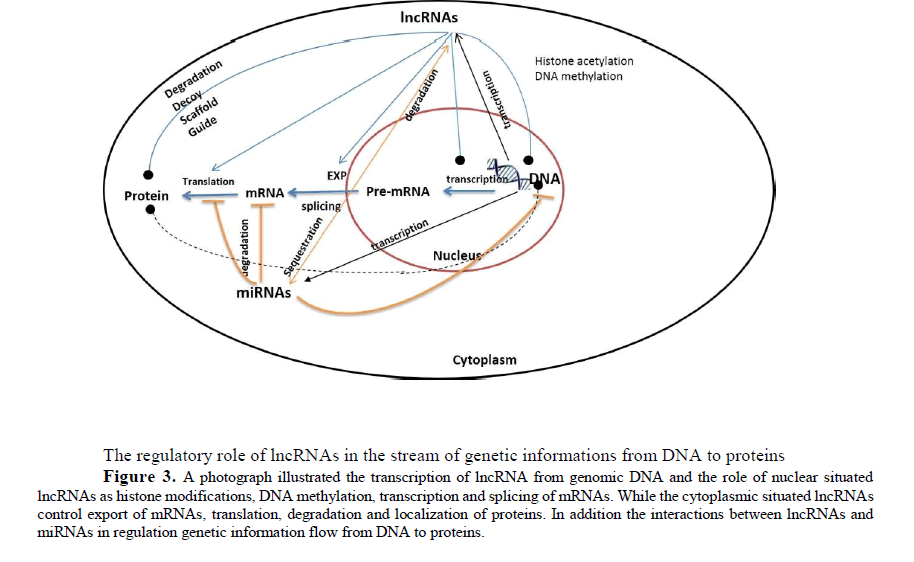
The regulatory role of lncRNAs in the stream of genetic informations from DNA to proteins
Figure 3: A photograph illustrated the transcription of lncRNA from genomic DNA and the role of nuclear situated lncRNAs as histone modifications, DNA methylation, transcription and splicing of mRNAs. While the cytoplasmic situated lncRNAs control export of mRNAs, translation, degradation and localization of proteins. In addition the interactions between lncRNAs and miRNAs in regulation genetic information flow from DNA to proteins.
The lncRNA pRNA interacts with DNA at the specific site for interaction DNMT3B to regulate rRNA transcription (Schmitz, K.M et al., 2010). In addition to that, PTENP1-asRNA alpha, inhibit transcription of PTEN coding gene by binding to DNMT3A (DNA methylase) at PTEN coding gene promoter and this complex is required for the deposition of repressive H3K27me3 chromatin marks at the PTEN promoter (Johnsson P et al., 2013). Moreover, PTENP1asRNA beta positively regulates PTENP1 mRNA post transcriptionally through PTENP1 sense (RNA-RNA interaction) and stabilizes it.
The nuclear enriched transcript 1 (NEAT1) nucleates the formation of paraspeckles at its transcription site by recruiting paraspeckle proteins (PSPC1, PSF/SFPQ and NONO/P54NRB) either directly or in a complex and its continued expression is required to maintain paraspeckles, identifying the dynamic nature of this nuclear body (Mao, Y.S et al., 2011, Clemson, CM et al., 2009). However, Neat1 Knocked out mouse were viable and fertile they have characteristic loss of paraspeckles (Nakagawa S et al., 2011).
Neat1 has also has epigenetic role as it repress gene expression by interaction with a number of chromatin binding protein/complexes in mouse embryonic stem cells including PRC1, PRC2, JARID1B, ESET and SUV39H1 (Guttman, M et al., 2011).
The lncRNA TERC is essential for the telomere complex assembly formation which maintains telomere length (Lustig, A.J et al., 2004). The interaction of H19 with MBD1 to form ribonucleoprotein complex that recruit histone lysine methyltrasnferases to suppress gene expression.
The lncRNA THRIL-hnRNPL interactions, regulates (TNF) α expression (Li, Z et al., 2014). These complexes regulate genes expressions at different levels which indicate the vital role of lncRNA interaction with different proteins to maintain cellular hemostasis
DNA double strand breaks (DSBs) occur in any given cell in the order of 10 to 50 per cell per day, depending on cell cycle and tissue (Vilenchik MM et al., 2003). DSBs are rare but it is highly toxic lesion requiring orchestrated and conserved machinery to prevent adverse consequences, such as cell death and cancer-causing genome structural mutations. DSBs trigger the DNA damage response (DDR) that directs a cell to repair the break, upon this DDR a number of lncRNAs-Protein complex plays a key role in the repair process. Among the already identified DSBs lncRNAs, the lncRNA PANDA that binds with transcription factor NF-YA. This complex lowers the activity of NFYA which interact with P53 the cell cycle key regulator and decreasing the activity of apoptotic genes (Di Agostino, S et al., 2006, Matuoka, K et al., 2000).
Another DSBs induced lncRNA, Gadd7 in Chinese Hamster ovary control G1/S checkpoint and cell growth by interacting with TAR DNA-binding protein (TDP-43) (Hollander MC et al., 1996). This Gadd7-TDP-43 complex interfered with the binding of TDP-43 to Cdk6 mRNA, leading to increasing turnover of Cdk6 mRNA resulted in abnormal cell cycle progression and incidence of senescence (Liu, X et al., 2012, Rader J et al., 2013) . While sequestering of Pumilio proteins, Pumilio-Fem3-biding factr (PUF) a negative regulator of gene expression, during chromosomal segregation by the lncRNA activated by DNA damage’’ NORAD maintains genomic stability. (Lee, S et al., 2016). By binding of NORAD to 3’ UTR of target mRNA through their PUMILIO homology domain and enhances deadenylation and decapping resulted in accelerated turnover and decreased translation(Zamore, PD et al., 1997, Miller MA et al., 2011)
In response to DSBs, the Ku80–Ku70 heterodimer associates with the broken ends, forming a clamp-like complex that recruits the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) to sites of damage. Additional protein factors, including Artemis, DNA ligase IV, XRCC4, and XLF assemble with the Ku80–Ku70–DNA-PK complex and promote processing and ligation of the broken ends (Helleday T et al., 2008, Mehta A et al., 2014). The lncRNA LINP1 was found to serve as a scaffold linking Ku80 and DNA-PKcs, thereby coordinating the NHEJ pathway and enhance the repair process (Zhang, Y et al., 2016).
The anchoring of the inactiveX chromosome, situated in the perinucleolar region during S phase maintained by ribonucleoprotein complex. The lncRNA Firre interacts with protein complex that includes CTCF, cohesion, YY1 and nucleophsmin not only to fix in active X chromosome but also to maintain its H3K27me3 (Yang F et al., 2015) (Yang F et al., 2015). Furthermore, XIST, a lncRNAs that is responsible for X chromosome inactivation in females, was shown to be declined in senescent cells (Umlauf DP et al., 2008, Sado T et al., 2013) but its specific function is still unknown.
On the other hand, the staffing of translational repressors (RCK and FMRP) by the human lncRNAp21suprres the translation of CTNNB and JUNB mRNAs (encoding β-catenin and JunB, respectively) (Yoon JH et al., 2012,). Although β-catenin and JunB are known to influence cell cycle proliferation and progression the exact role of lncRNA-p21 is still obscure (Konishi N et al., 2008, Ye, X et al., 2007).
The interaction of different a number lncRNAs with the well-known repressor proteins PRC1 and PRC2 acts as transcriptional repressors for different genomic loci. For example, ANRIL (Antisense noncoding RNA in the INK4 locus), interacts with both SUZ12 a component of PRC2 and CBX7, a component of PRC1, to accelerate epigenetic silencing of the CDKN2A/CDKN2B loci. Further, its down regulation increases expression of the cell cycle inhibitor P14,P15 and P16 (Kotake Y et al., 2011, Yap KL et al., 2010). In addition to that, the lncRNA MIR31HG interacts with both complexes PRC1 and PRC2 to repress P16INK4A expression (Montes M et al., 2015).
The oncogenic lncRNA Focally amplified on chromosome 1 (FAL-1) interacts with and stabilizes BMI1, a PRC1 component, resulting in transcriptional repression of p21 (Hu, X et al.,2014). While the senescence associated lncRNA SALNR interacts with NF90, a nuclear protein that binds and prevents the biogenesis of senescence-associated miRNAs (Tominaga-Yamanaka K et al., 2012) and its over-expression of inhibits the localization of NF90 to nucleoli, which delays the onset of senescence (Wu, C.L et al., 2015).
Cytoplasmic lncRNAs
The molecular mechanisms for lncRNAs located in the cytoplasm have been less investigated. They may function as translation regulators via base pairing with their target mRNAs [60, 70] or may increase or decrease mRNA stability to affect protein expression levels (Faghihi MA et al., 2008, Kretz M et al., 2013). Another function for cytoplasmic lncRNAs is to regulate ubiquitination process or they control the passage of proteins or other RNA between the cytoplasm and the nucleus Figure (3).
The lncRNA GAS5, acts a s a decoy of glucocorticoid receptor (GR) and prevent its mobilization from the cytosol to the nucleus thus repress GR mediated gene expression (Mourtada-Maarabouni, M et al., 2009, Lee, S.Y et al., 2012).
The importance of P53 as a tumor suppressor is well exemplified by the estimated that it can drive around 50% of all cancers. It is subjected to exquisite control via multiple mechanisms involving array of interaction partners. One of such partners, the lncRNA MEG3 (Maternally expressed gene 3) induces accumulation of p53 protein, directly via RNA-protein association or indirectly by lowering MDM2 expression levels (Baldassarre, A et al., 2012, Tasdemir E et al., 2008). On the other hand, the highly up regulated lncRNA 7SL, interacts with P53 mRNA and suppress P53 translation.
Another P53 partner, The RNA binding protein HuR can displace 7SL and enhance P53 translation. According to this competitive interaction, they can control cell cycle. As silencing of 7SL increased HuR binding to TP53 mRNA and promoted p53 translation, in turn enhancing cell cycle arrest and senescence (Abdelmohsen, K et al., 2014).
The steady state of protein abundance in the cell is controlled by protein synthesis and degradation. Some lncRNA modulated protein levels indirectly by influencing the available pool of microRNAs and theryby affecting mRNA turnover and translation. For instance the lncRNA linc-MD1 and lincRNA-ROR acts as a decoy for miRNAs that usually suppress the translation or stability of other mRNAs (Wang Y et al., 2013, Cesana M et al., 2011). However, some lncRNAs interacts directly with mRNAs to enhance or suppress their translations or with proteins to modulate their half-life time.
The lncRNA (AS Uchl1), enhances translation of UCHL1 (ubiquitin carbosy-terminal hydroxylase L1) through an imbedded SINES B2 present in AS Uchl1. UCHL1 is also, involved in brain development and age related neurodegenerative pathologies as Parkinson’s disease and its overexpression was associated senescence induction, likely due to increase production of p14ARF, p27KIPI and decreased production of MDM2 levels [80].
The senescence up regulated lncRNA HOTAIR, enhances ubiquitination and subsequent degradation of Ataxin-1 (ATXN1) and Snurportin-1 (SNUPN) by acting as a scaffold for the E3 ubiquitin ligases Mex-3 RNA binding family member B (MEX3B) and DAZ interacting zinc finger protein (DZIP3) and their respective substrates [81] while, HuR promotes HOTAIR decay, and loss of HuR during senescence may contribute to HOTAIR stabilization and subsequent up-regulation (Wang,W et al., 2001).
Notably, LncRNAs expression is strikingly cell type or tissue restricted and, in many cases, is even primate specific. Investigations of lncRNAs have demonstrated that they can serve as scaffolds or guides regulating protein-protein or protein-DNA interactions, as decoys that bind proteins or microRNAs (miRNAs), or as enhancers of gene expression when transcribed within enhancer regions or their neighboring loci.
Closing remarks
Thousands of lncRNA are expressed in human cells. The functions of most on these lncRNAs are unknown, but their expression can be highly cell type or tissue specific and many were deregulated either in physiological or pathological conditions to maintain cellular homeostasis. Further, the implication of majority of lncRNAs in diseases conditions as cancer, apoptosis, neurodegenerative diseases and senescence. As the already characterized lncRNAs identified in pathological conditions does these transcripts are functional in physiological conditions or not? Or does these lncRNAs are abundant in normal cell state?
With the advent of sensitive high throughput genomic technologies, as microarray and next generation sequencing (NGS) a large fraction of the mammalian genome is likely to be transcribed, the characteristic and functions of overwhelming majority of lncRNA are cruuently unknown. Some of them are cytoplasmic others are nuclear, some are highly expressed others are barely detected. And functional studies indicate important role of several lncRNA, the majority of them a wait for further verification. In the last decade, the field of lncRNA becomes one of the fastest moving areas of research, which makes this the exciting time for the study on ncRNA realm.
Long non-coding RNAs (lncRNAs) are associated to a plethora of cellular functions, most of which require the interaction with one or more RNA-binding proteins (RBPs); similarly, RBPs are often able to bind a large number of different RNAs. The currently available knowledge is already drawing an intricate network of interactions, whose deregulation is frequently associated to pathological states. Several different techniques were developed in the past years to obtain protein–RNA binding data in a high-throughput fashion. In parallel, in silico inference methods were developed for the accurate computational prediction of the interaction of RBP–lncRNA pairs. The field is growing rapidly, and it is foreseeable that in the near future, the protein–lncRNA interaction network will rise, offering essential clues for a better understanding of lncRNA cellular mechanisms and their disease-associated perturbations.
However, only not more than a handful of lncRNA were identified, identification the interactions of the proteins/mRNAs that interact with lncRNA can the possible guide to predict functions of these lncRNA. At the present time we still have some limitations to identify the detailed functions of different lncRNA as lack of the exact protocol that can be applied for all ncRNAs, lack of conserved nature of most of lncRNA. By advancing in these areas of knowledge, we expect to gain a deeper molecular understanding of the exact role of lncRNA and the aura of lncRNA mystery will be gradually unveiled. At the same time, we can answer the following questions, does the interaction between lncRNAs and proteins is temporarily or controlled by the half life time of either lncRNA or protein? Is lncRNA acts as cellular thermostat and give the alarm when the cell hemostasis changed either under physiological or pathological alterations? Is the nature of this interaction is a kind of marriage between both? The possibility of the use of lncRNAs for therapeutic purposes through identification of their dynamic molecular functions at different cellular levels.
Disclosure Statement
The authors declare that they have no conflicts of interests.
Acknowledgments
We are grateful for the assistance of Dr. Louise C. Abbott in editing this manuscript as well as the anonymous reviewers whose comments will enrich our manuscript. A.R.G. is a recipient of Chinese Scholarship Council (CSC), W.A and M.A is a recipient of CAS-TWAS President's PhD Fellowship
About the Authors
References
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215-33. https://doi.org/10.1016/j.cell.2009.01.002
- Gong C and Maquat LE (2011) lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature 470(7333): 284-288.
- Payre F and Desplan C (2016) RNA. Small peptides control heart activity. Science 351(6270): 226-7. https://doi.org/10.1126/science.aad9873
- Li JH., et al., (2014) starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 42(Database issue):√?¬† D92-97. https://doi.org/10.1093/nar/gkt1248
- Paraskevopoulou MD, et al (2013) DIANA-LncBase: experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res,. 41(Database issue): D239-45. https://doi.org/10.1093/nar/gks1246
- Powers ET et al., (2009) Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem 78: 959-91. https://doi.org/10.1146/annurev.biochem.052308.114844
- Balch WE., et al., (2008) Adapting proteostasis for disease intervention. Science 319(5865): 916-919. https://doi.org/10.1126/science.1141448
- Ghanam AR, Shengwei Ke QX, Muhammad Azhar, Qingyu Cheng, Xiaoyuan Song, et al., (2017) Shining the Light on Senescence Associated LncRNAs. A &D 8: 149. https://doi.org/10.14336/ad.2016.0810
- Hartl FU, Bracher A and Hayer-Hartl M (2011) Molecular chaperones in protein folding and proteostasis. Nature 475(7356): 324-332. https://doi.org/10.1038/nature10317
- Koga HS, Kaushik and Cuervo AM (2011) Protein homeostasis and aging: The importance of exquisite quality control. Ageing Res Rev10: 205-15. https://doi.org/10.1016/j.arr.2010.02.001
- Deo RC, et al., (1999) Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell 98: 835-45. https://doi.org/10.1016/s0092-8674(00)81517-2
- Wang X and Tanaka Hall TM (2001) Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat Struct Biol 8: 141-5.
- Shamoo Y, et al (1997) Crystal structure of the two RNA binding domains of human hnRNP A1 at 1.75 A resolution. Nat Struct Biol 4: 215-22. https://doi.org/10.1038/nsb0397-215
- Mazza C, et al., Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J, 2002. 21(20): p. 5548-57. https://doi.org/10.1093/emboj/cdf538
- Liker E, et al., (2000) The structure of the mRNA export factor TAP reveals a cis arrangement of a non-canonical RNP domain and an LRR domain. EMBO J 19: 5587-98. https://doi.org/10.1093/emboj/19.21.5587
- Conte MR, et al., (2000) Structure of tandem RNA recognition motifs from polypyrimidine tract binding protein reveals novel features of the RRM fold. EMBO J 19: 3132-41. https://doi.org/10.1093/emboj/19.12.3132
- Perez Canadillas JM and Varani G (2003) Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. EMBO J 22: 2821-30. https://doi.org/10.1093/emboj/cdg259
- Inoue M, et al., (1997) A characteristic arrangement of aromatic amino acid residues in the solution structure of the amino-terminal RNA-binding domain of Drosophila sex-lethal. J Mol Biol 272: 82-94. https://doi.org/10.1006/jmbi.1997.1213
- Varani, L., et al., (2000) The NMR structure of the 38 kDa U1A protein - PIE RNA complex reveals the basis of cooperativity in regulation of polyadenylation by human U1A protein. Nat Struct Biol 7: 329-35. https://doi.org/10.2210/pdb1dz5/pdb
- Jacks A, et al., (2003) Structure of the C-terminal domain of human La protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element. Structure 11: 833-43. https://doi.org/10.1016/s0969-2126(03)00121-7
- Simpson PJ, et al., (2004) Structure and RNA interactions of the N-terminal RRM domains of PTB. Structure 12: 1631-43. https://doi.org/10.1016/j.str.2004.07.008
- Oubridge C, et al., (1994) Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 372(6505): 432-8. https://doi.org/10.1038/372432a0
- Lewis HA., et al., (2000) Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell 100: 323-32. https://doi.org/10.1016/s0092-8674(00)80668-6
- Ramos A., et al., (2000) RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J 19: 997-1009.
- Lu D, Searles MA, and Klug A (2003) Crystal structure of a zinc-finger-RNA complex reveals two modes of molecular recognition. Nature 426(6962): 96-100. https://doi.org/10.2210/pdb1un6/pdb
- Hudson BP, et al., (2004) Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol 11: 257-64. https://doi.org/10.1038/nsmb738
- Frazao C., et al., (2006) Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature 443(7107): 110-114. https://doi.org/10.1038/nature05080
- Ma JB, et al., (2005) Structural basis for 5'-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434(7033): 666-70. https://doi.org/10.2210/pdb1ytu/pdb
- Antson AA, et al., (1999) Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature 401(6750): 235-42. https://doi.org/10.2210/pdb3zte/pdb
- Rinn, JL and Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81: 145-66.
- Guttman M, and Rinn JL (2012) Modular regulatory principles of large non-coding RNAs. Nature 482(7385): 339-46. https://doi.org/10.1038/nature10887
- Schmitz KM., et al., (2010) Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 24: 2264-9. https://doi.org/10.1101/gad.590910
- Johnsson P, et al., (2013) A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol 20: 440-446. https://doi.org/10.1038/nsmb.2516
- Mao YS, et al., (2011) Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol 13: 95-101. https://doi.org/10.1038/ncb2140
- Sunwoo H, et al., (2009) MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19: 347-59. https://doi.org/10.1101/gr.087775.108
- Sasaki YT, et al., (2009) MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci USA 106: 2525-30. https://doi.org/10.1073/pnas.0807899106
- Clemson CM, et al., (2009) An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33: 717-26. https://doi.org/10.1016/j.molcel.2009.01.026
- Nakagawa, S., et al., (2011) Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol, 193: 31-39. https://doi.org/10.1083/jcb.201011110
- Guttman, M., et al., (2011) lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature, 477(7364): p. 295-300.
- Lustig AJ., (2004) Telomerase RNA: a flexible RNA scaffold for telomerase biosynthesis. Curr Biol 14: R565-7. https://doi.org/10.1016/j.cub.2004.07.013
- Li Z., et al., (2014) The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci USA 111: 1002-7. https://doi.org/10.1073/pnas.1313768111
- Vilenchik MM and Knudson AG (2003) Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc Natl Acad Sci USA 100: 12871-6. https://doi.org/10.1073/pnas.2135498100
- Di Agostino S., et al., (2006) Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell 10: 191-202. https://doi.org/10.1016/j.ccr.2006.08.013
- Matuoka K and Chen KY (2000) Possible role of subunit A of nuclear factor Y (NF-YA) in normal human diploid fibroblasts during senescence. Biogerontology 1: 261-71.
- Hollander MC, Alamo I, and Fornace AJ (1996) A novel DNA damage-inducible transcript, gadd7, inhibits cell growth, but lacks a protein product. Nucleic Acids Res 24: 1589-93. https://doi.org/10.1093/nar/24.9.1589
- Liu X., et al., (2012) Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. EMBO J 31: √?¬†4415-27. https://doi.org/10.1038/emboj.2012.292
- Rader J, et al., (2013) Dual CDK4/CDK6 inhibition induces cell-cycle arrest and senescence in neuroblastoma. Clin Cancer Res 19: 6173-82. https://doi.org/10.1158/1078-0432.ccr-13-1675
- Lee S, et al., (2016) Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell 164: 69-80. https://doi.org/10.1016/j.cell.2015.12.017
- Zamore PD, Williamson JR, Lehmann R (1997) The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA 3: 1421-33.
- Miller MA, and Olivas WM (2011) Roles of Puf proteins in mRNA degradation and translation. Wiley Interdiscip Rev RNA 2: 471-92. https://doi.org/10.1002/wrna.69
- Helleday, T., et al., DNA repair pathways as targets for cancer therapy. Nat Rev Cancer, 2008. 8(3): p. 193-204.
- Jackson SP. and Bartek J (2009) The DNA-damage response in human biology and disease. Nature 461(7267): 1071-8. https://doi.org/10.1038/nature08467
- Lieber MR (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79: 181-211. https://doi.org/10.1146/annurev.biochem.052308.093131
- Goldstein M, Kastan MB (2015) The DNA damage response: implications for tumor responses to radiation and chemotherapy. Annu Rev Med 66: 129-43. https://doi.org/10.1146/annurev-med-081313-121208
- Mehta A and Haber JE (2014) Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol 6: a016428. https://doi.org/10.1101/cshperspect.a016428
- Zhang Y., et al., (2016) Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer. Nat Struct Mol Biol, https://doi.org/10.1038/nsmb.3211
- Yang, F., et al., (2015) The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol 16: 52. https://doi.org/10.1186/s13059-015-0618-0
- Umlauf D, Fraser P, and Nagano T (2008) The role of long non-coding RNAs in chromatin structure and gene regulation: variations on a theme. Biol Chem 389: 323-31. https://doi.org/10.1515/bc.2008.047
- Sado T and Brockdorff N (2013) Advances in understanding chromosome silencing by the long non-coding RNA Xist. Philos Trans R Soc Lond B Biol Sci 368(1609): 20110325. https://doi.org/10.1098/rstb.2011.0325
- Yoon, JH, et al., (2012) LincRNA-p21 suppresses target mRNA translation. Mol Cell 47: 648-55. https://doi.org/10.1016/j.molcel.2013.04.008
- Konishi N, et al., (2008) Function of JunB in transient amplifying cell senescence and progression of human prostate cancer. Clin Cancer Res 14: 4408-16. https://doi.org/10.1158/1078-0432.ccr-07-4120
- Marchand A, et al (2011) The Wnt/beta-catenin pathway is activated during advanced arterial aging in humans. Aging Cell 10: 220-32. https://doi.org/10.1111/j.1474-9726.2010.00661.x
- Ye X., et al., (2007) Downregulation of Wnt signaling is a trigger for formation of facultative heterochromatin and onset of cell senescence in primary human cells. Mol Cell 27: 183-96. https://doi.org/10.1016/j.molcel.2007.05.034
- Kotake Y., et al., (2011) Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 30:1956-62. https://doi.org/10.1038/onc.2010.568
- Yap, K.L., et al., (2010) Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 38: 662-74. https://doi.org/10.1016/j.molcel.2010.03.021
- Montes M., et al., (2015) The lncRNA MIR31HG regulates p16(INK4A) expression to modulate senescence. Nat Commun 6: 6967. https://doi.org/10.1038/ncomms7967
- Hu X., et al., (2014) A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell 26: 344-57. https://doi.org/10.1016/j.ccr.2014.07.009
- Tominaga-Yamanaka K, et al., (2012) NF90 coordinately represses the senescence-associated secretory phenotype. Aging (Albany NY) 4: 695-708. https://doi.org/10.18632/aging.100497
- Wu CL et al., (2015) Senescence-associated Long Non-coding RNA (SALNR) Delays Oncogene-induced Senescence through NF90 Regulation. J Biol Chem 290: 30175-92. https://doi.org/10.1074/jbc.m115.661785
- Carrieri C, et al., (2012) Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491(7424): 454-7. https://doi.org/10.1038/nature11508
- Faghihi MA, et al., (2008) Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med 14:723-30. https://doi.org/10.1038/nm1784
- Kretz M, et al., (2013) Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493(7431): 231-235. https://doi.org/10.1038/nature11661
- Mourtada-Maarabouni M, et al., (2009) GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 28:195-208. https://doi.org/10.1038/onc.2008.373
- Lee SY, et al., (2012) Decreased levels of nuclear glucocorticoid receptor protein in the hippocampus of aged Long-Evans rats with cognitive impairment. Brain Res 1478: 48-54. https://doi.org/10.1016/j.brainres.2012.08.035
- Baldassarre A and Masotti A (2012) Long non-coding RNAs and p53 regulation. Int J Mol Sci 13:√?¬† 16708-17. https://doi.org/10.3390/ijms131216708
- Tasdemir E, et al., (2008) A dual role of p53 in the control of autophagy. Autophagy 4: 810-814. https://doi.org/10.4161/auto.6486
- Abdelmohsen K, et al., (2014) 7SL RNA represses p53 translation by competing with HuR. Nucleic Acids Res, 42: 10099-111. https://doi.org/10.1093/nar/gku686
- Wang Y, et al., (2013) Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell, 25: 69-80. https://doi.org/10.1016/j.devcel.2013.03.002
- Cesana M, et al., (2011) A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147: 358-69. https://doi.org/10.1016/j.cell.2011.10.031
- Ummanni R et al., (2011) Ubiquitin carboxyl-terminal hydrolase 1 (UCHL1) is a potential tumour suppressor in prostate cancer and is frequently silenced by promoter methylation. Mol Cancer 10: 129. https://doi.org/10.1186/1476-4598-10-129
- Yoon JH, et al., (2013) Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun 4: 2939. https://doi.org/10.1038/ncomms3939
- Wang W, et al., (2001) Loss of HuR is linked to reduced expression of proliferative genes during replicative senescence. Mol Cell Biol 21: 5889-98. https://doi.org/10.1128/mcb.21.17.5889-5898.2001
Keywords:
Download:
Full PDF- Share This