Possible association of proinflammatory cytokine IL-19 gene polymorphism with psoriasis
Received: December 03, 2018
Accepted: January 01, 1970
Published: January 05, 2019
Genet.Mol.Res. 18(1):
Introduction
Psoriasis may be considered a result of dysregulation of the cytokines which is responsible for most of the clinical features of psoriasis like hyperproliferation of keratinocytes, inflammation and increased neovascularization. Understanding the genetic risk may help to determine which genes control which aspects of psoriasis it may alsoidentify potential novel therapeutic targets for management of psoriasis. Current management of psoriasis is based on treat to target where cytokines that play a central role in the disease development are being targeted (Gudonsson J, Johnston A, Sigmundsdottir H, Valdimarsson H, 2004). Psoriasis effects 0.44 to 2.80% of population inIndia (Finch CE and Crimmins EM, 2004; Wani A, Ganai BA, Akhtar T, Narang T, et al., 2017; Dogra S and Yadav S, 2010). IL-19 is secreted as monomer by activated monocytes and to a lesser extent by B cells produce (Gallagher G, Dickensheets H, Eskdale J, Izotova LS, et al., 2000; Wolk K, Kunz S, Asadullah K, Sabat R, 2002). IL-19 having seven helices able to bind its receptor which act through the IL-20R (1) /IL-20R (2) heterodimer involved in triggering proliferation and abnormal keratinocyte differentiation (Gallagher G, Dickensheets H, Eskdale J, Izotova L, et al., 2000; Koks S, Kingo K, Rätsep R, Karelson M, 2004). Binding of IL-19 to its receptors leads to rapid activation of the transcription factor STAT3 (activators of transcription 3 and signal transducers (Dumoutier L, Van Roost E, Colau, D, Renauld JC, 2000). Gene expression of IL-19 is increased in inflammatory skin diseases particularly in psoriatic lesional skin compared to non-lesional psoriatic skin and has been marked as biomarker for activity of psoriatic diseases (Witte E, Kokolakis G, Witte K, Philipp S Wittig, 2014). In vitro analysis demonstrates that IL-19 stimulates the production of IL-6 and tumor necrosis factor-alpha (TNF-α) from monocytes signifying that IL-19 might be a proinflammatory cytokine (Liao YC, Liang WG, Chen FW, Hsu JH, 2002). On distinction inflammation is now considered to be an important risk factor for aging (Finch CE and Crimmins EM, 2004; Franceschi C, Bonafè M, Valensin S, Olivieri F, 2000). In the sight of the above results, IL-19 appears to be the contender gene for psoriasis understanding. The present study was designed to investigate the role of IL- 19 polymorphism, haplotype analysis as a risk factor for the development of psoriasis in north India. To our knowledge no polymorphism study of IL-19 genes has been reported till now in north Indian population.
Material and Methods
Subject recruitment
This hospital based case-control study was conducted after approval by Institutional ethical committee. The subjects were included only after they willingly decided to become part of the study, and filled the consent form. This study includes 200 cases of psoriatic that were diagnosed by expert dermatologists. Doubtful cases were recruited only after histo-pathological confirmation of diagnosis. Gender and geographically matched healthy subjects were included as controls in the study. Freshly diagnosed psoriatic patients (prior to treatment) and patients which have undergone off treatment for three weeks were included while as Psoriatic patients who were suffering from any other autoimmune disorders, acute or chronic infections and malignancies were excluded A complete history and demographic profile of the patient was noted on a prescribed performa, psoriasis severity was also assessed in all the recruited patients. Psoriasis Area and Severity Index (PASI) is widely used tool for the measurement of severity of psoriasis. PASI score was calculated as described by Langle et al. (Langley RG and Ellis CN, 2004).
Blood sampling
Five ml of venous blood was taken from each subject and was divided into two portions. 2 ml was taken in sterile EDTA coated vials for Genomic DNA Extraction and the remaining was centrifuged at 4000 rpm for 5 mins. Serum separated was stored at -80°C till further analysis.
Genomic DNA extraction
Genomic DNA was isolated from the blood samples by using Phenol-Chloroform method (Sambrook J and Russell DW, 2006) and the isolated DNA was stored at -20°C for future use.
Genotyping of IL-19
Novel tetra-primer ARMS-PCR method was applied for genotyping of polymorphisms of IL-19 (Kingo K, Koks S, Nikopensius T, Silm H, 2004; Wani A, Ganai BA, Akhtar T, Narang T, 2018). The tetra-primer ARMS-PCR method employs two primer pairs to amplify, respectively, the two different alleles of an SNP in a single PCR reaction. Either the allele-specific amplicons are generated using one allele-specific inner primer and one non-allele-specific outer primer. Outer primers are also used to generate a non-allele-specific control product. Sets of four primers, specific to each SNP, were designed by Web-based allele specific primer designing tool (WASP). Each PCR reaction was carried out in a total volume of 10 μl containing 100 ng of template DNA, 20 pmol of each inner primer, 20 pmol of each outer primer, and optimized concentrations of master mix. To increase the specificity of a PCR reaction we applied touchdown cycles: initial denaturation at 95°C for 2 min followed by 10 cycles of 1 min denaturation at 95°C, annealing at 10°C higher than annealing temperature for 1 min (decreasing by 1°C per cycle) and extension at 72°C for 1 min.
Statistical Analysis
Categorical variables were set for presenting and calculating numbers and percentages for different variants of IL-19. Conditional logistic regression models were used to calculate odds ratios (ORs) and corresponding 95% confidence intervals (CIs) to assess the association of various polymorphisms of IL-19 with Psoriasis. The adjustment was made for known risk factors like age, family history and smoking. All statistical analysis was done using SNP Stat software. Two-sided p<0.05 was considered as statistically significant.
Results
Two hundred confirmed psoriatic cases and an equal number of healthy, age and gender matched controls were recruited in this study. After analyzing the data out of 200 cases recruited in the present study, 130 (65%) were males and 70 (35%) were females. It was found that age ranged from 18-70 years with mean 38.61 ± 13.71 in the psoriatic patients while in controls, age ranged from 19-65 years with mean value of 36.695 ± 11.47. Thirty two (16%) psoriatic patients had positive family history for psoriasis. majority of the patients (57.5%) were from rural areas whiles 42.5% were from urban areas. Body Mass Index (BMI) of psoriatic patients and controls was 26.94 ± 4.17 Kg/m2 and 24.80 ± 4.28 Kg/m2 respectively. The mean value of PASI in psoriatic patients was (10.654 ± 9.09) (Table 1).
| Characteristics | Cases (%) | Controls (%) |
|---|---|---|
| Age: X ± SD | 38.61 ± 13.71 years | 36.695 ± 11.47 Year |
| Sex Male Female |
130 (65.00%) | 130 (65.00%) |
| 70 (35.00%) | 70 (35.00%) | |
| Family history of psoriasis | 32 (16.00%) | ----- |
| Disease duration ± SD | 9.88 ± 8.19 years | |
| Dwelling Rural Urban |
115 (57.50%) 85 (42.50%) |
115 (57.50%) 85 (42.50%) |
| Smoking Smoker Non-Smoker |
30 (15%) 170 (85%) |
09 (4.5% 191 (95.5%) |
| BMI (kg/m2) X ± SD | 26.94 ± 4.17 | 24.80 ± 4.28 |
| PASI: X ± SD | 10.654 ± 9.09 | ----- |
Table 1. Characteristics of study group.
The allele and genotype frequencies of IL-19-42232 C/A (rs2243188), IL-19-38386 T/C (rs2073186) &IL-19 -35402 G/C (rs2243158), in patients and controls are summarized in Tables 2, 3 and 4. Genotype distributions for the three analyzed IL-19 gene polymorphisms were found in Hardy–Weinberg equilibrium.
| Variables | Genotype | Cases n (%) |
Controls n (%) |
Un adjusted OR (95% CI) | Adjusted *OR (95% CI) |
P value |
|---|---|---|---|---|---|---|
| Il 19 -42232 C/A (rs2243188) |
C/C | 87 (43.50) | 72 (36.00) | 1 | 1 | |
| C/A | 96 (48.00) | 106 (53.00) | 0.75 (0.49 – 1.14) | 0.71 (0.44 – 1.14) | 0.21 | |
| A/A | 17 (8.50) | 22 (11.00) | 0.641 (0.32 – 1.30) | 0.59 (0.25 – 1.40) | 0.26 | |
| Dominant | C/C | 87 (43.50) | 72 (36.00) | 1.00 | 1 | 0.11 |
| C/A +A/A | 113 (56.50) | 128 (64.00) | 0.73 (0.49 – 1.09) | 0.69 (0.43 - 1.09) | ||
| Recessive | C/C+A/A | 183 (91.50) | 178 (89.00) | 1.00 | 1.00 | 0.42 |
| A/A | 17 (8.50) | 22 (11.00) | 0.75 (0.39 – 1.46) | 0.71 (0.31 - 1.63) | ||
| Overdominant | C/C+A/A | 104 (52.00) | 94 (47) | 1.00 | 1 | 0.27 |
| C/A | 96 (48.00) | 106 (53.00) | 0.82 (0.55 – 1.21) | 0.77 (0.49 - 1.22) | ||
| Allele | C A |
270 (67.50) 130 (32.50) |
250 ( 62.50) 150 (37.50) |
1 0.80 (0.59 – 1.07) |
0.15 | |
n=Number of Individuals, Statistically significant p<0.05
*Adjusted OR (95%CI) were obtained when OR was adjusted for age, smoking and family history
Table 2. Distribution of genotypes and allele frequency of IL-19 -42232 C/A (rs2243188) polymorphism in psoriasis cases and controls.
| Genotype | Cases n (%) |
Controls n (%) |
Un adjusted OR (95% CI) | Adjusted *OR (95% CI) |
P value | |
|---|---|---|---|---|---|---|
| Il 19 – 38386 C/T (rs 2073186) |
C/C | 61 (30.50) | 69 (34.50) | 1 | 1 | |
| T/C | 81 (40.40) | 86 (43.00) | 1.07 (0.67 – 1.69) | 1.22 (0.71 – 2.08) | 0.88 | |
| T/T | 58 (29.00) | 45 (22.50) | 1.46 (0.87 – 2.45) | 1.28 (0.70 – 2.35) | 0.68 | |
| Dominant | C/C | 61 (30.50) | 69 (34.50) | 1.00 | 1 | 0.39 |
| T/C +T/T | 139 (69.50) | 131 (65.50) | 1.20 (0.79 - 1.82) | 1.24 (0.76 – 2.03) | ||
| Recessive | C/C+T/C | 142 (71.00) | 155 (77.50) | 1.00 | 1.00 | 0.62 |
| T/T | 58 (29.00) | 45 (22.50) | 1.41 (0.90 – 2.21) | 1.14 (0.68 – 1.91) | ||
| Over-dominant | C/C+T/T | 119 (59.50) | 114 (57.00) | 1.00 | 1 | 0.73 |
| T/C | 81 (40.50) | 86 (43.00) | 0.90 (0.61- 1.34) | 1.08 (0.69 – 1.71) | ||
| Allele | C T |
203 (50.75) 197 (49.25) |
224 (56.00) 176 (44.00) |
1 1.23 (0.93 - 1.63) |
0.156 | |
n=Number of Individuals, Statistically Significant p<0.05
*Adjusted OR (95%CI) were obtained when OR was adjusted for age, smoking and family history
Table 3. Distribution of genotypes and allele Frequency of IL-19 -38386 C/T (rs 2073186) polymorphism in psoriasis cases and controls
| Genotype | Cases n (%) |
Controls n (%) |
Un adjusted OR (95% CI) | Adjusted *OR (95% CI) |
P value | |
|---|---|---|---|---|---|---|
| Il 19 – 35402 G/C (rs 2243158) | G/G | 91 (45.50) | 144 (72.00) | 1 | 1 | |
| G/C | 53 (26.50) | 13 (6.50) | 6.45 (3.33 - 12.49) | 7.13 (3.36 – 15.12) | 0.0001 | |
| C/C | 56 (28.00) | 43 (21.50) | 2.06 (1.28 - 3.32) | 2.05 (1.19 – 3.55) | 0.0001 | |
| Dominant | G/G | 91 (45.50) | 144 (72.00) | 1.00 | 1 | 0.0001 |
| G/C +C/C | 109 (54.50) | 56 (28.00) | 3.08 (2.03 - 4.67) | 3.12 (1.92 – 5.06) | ||
| Recessive | G/G+G/C | 144 (72.00) | 157 (78.5) | 1.00 | 1.00 | 0.26 |
| C/C | 56 (28.00) | 43 (21.50) | 1.42 (0.90 - 2.24) | 1.34 (0.80 – 2.25) | ||
| Overdominant | G/C+C/C | 147 (73.50) | 187 (93.50) | 1.00 | 1 | 0.0001 |
| G/C | 53 (26.50) | 13 (6.50) | 5.19 (2.72 - 9.87) | 5.56 (2.70 – 11.46) | ||
| Allele | G C |
235 (58.75) 165 (41.25) |
301 (75.25) 99 (24.75) |
1 2.134 (1.57 - 2.88) |
0.0001 | |
n=Number of Individuals Statistically Significant p<0.05
*Adjusted OR (95%CI) were obtained when OR was adjusted for age, smoking and family history.
Table 4. Distribution of genotypes and allele Frequency of IL-19-3540 G/C (rs2243158) polymorphism in psoriasis cases and controls
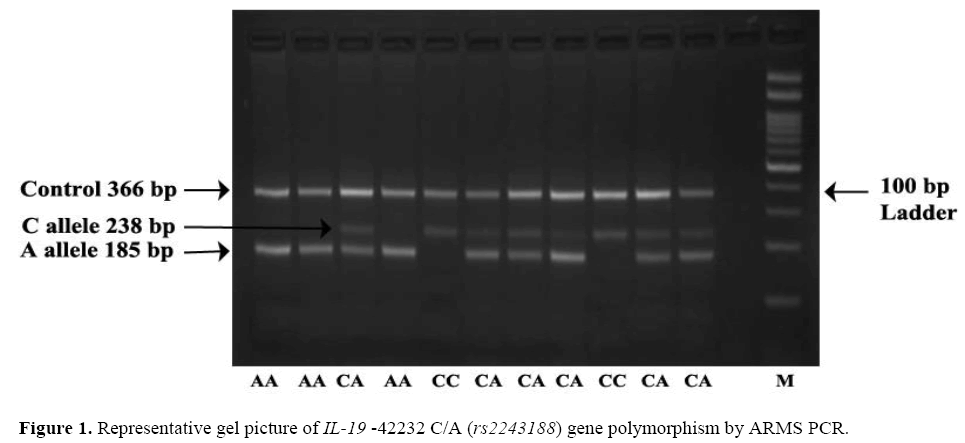
The frequency of minor allele A at position –42232 C/A ( rs2243188) was found higher in controls 37.50% (150/400) as compared to cases 32.50% (130/400) OR=0.80; (95% CI=0.59 – 1.07) the difference in allelic frequency was found statistically non-significant. Observation shows that AA genotype was higher in controls 11% (22/200) than cases 8.50% (17/200) adjusted* OR=0.59; (95% CI=0.25 – 1.40), the difference was found to bestatistically non-significant which leads to confirmation that –42232 AA genotype is not associated with risk of psoriasis. The dominant, recessive and overdominant models does not show any association as a risk factor for psoriasis. Representative gel picture of IL-19 –42232 C/A ( rs2243188) gene polymorphism by ARMS PCR (Figure 1).
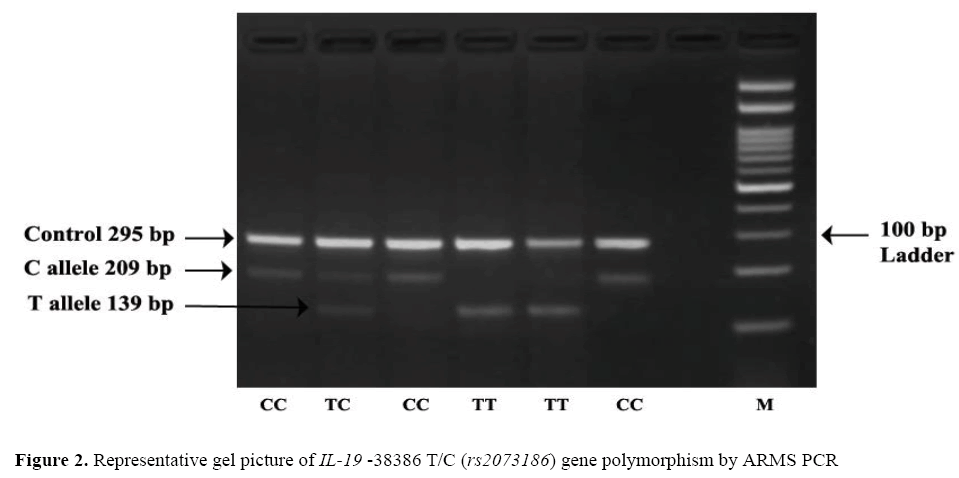
The frequency of minor allele T at position-38386 T/C (rs2073186) was found higher in cases 49.25% (197/400) as compared to that of controls 44% (176/400) OR=1.23; (95% CI=0.93- 1.63) the difference wasstatistically non-significant (p=0.156). Observation shows that TT genotype was higher in cases 29.00% (58/200 than controls 22.5% (45/200) adjusted* OR=1.28; (95% CI=0.70 – 2.35) the difference was statistically non-significant (p=0.68), which leads confirmation that –38386TT genotype was not associated with risk of psoriasis. The dominant, recessive and overdominant models does not show any association as a risk factor for psoriasis as the results were statisticallynon significant. Representative gel picture of IL-19 –38386 T/C (rs2073186) gene polymorphism by ARMS PCR. ( Figure 2).
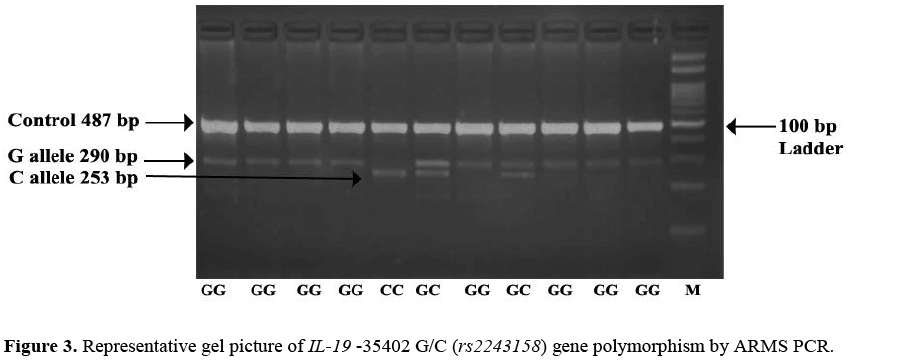
The minor allele C frequency at position -35402 G/C (rs2243158) was found higher in cases 41.25% (165/400) as compared to that of controls 24.75% (99/400) OR=2.134; (95% CI=1.57 – 2.88) the difference was statistically significant. Observation shows that CC genotype was higher in cases 28.00% (56/200) than controls 21.5% (43/200) adjusted* OR=2.05; (95% CI=1.19 – 3.55) the difference wasstatistically significant which leads to the confirmation that -35402 C/C genotype is associated with increased risk of psoriasis. The dominantadjusted* OR=3.12; (95% CI=1.92 – 5.06) and overdominantadjusted* OR=5.56; (95% CI=2.70 – 11.46) model shows association of genotypes as a increased risk factor for psoriasis as the results arestatistically significant wher as recessive model does not show any significant association with psoriasis. Representative gel picture of IL-19 -35402 G/C (rs2243158) gene polymorphism by ARMS PCR. (Figure 3).
Haplotype frequencies of psoriasis patients and controls are summarized in Table 5 The pair wise LD Matrix demonstrated that the nearly complete L.D. (D >0 <1) (D, between 0.50 to 0.70) existed between the polymorphism at position –42232, -3836 and -35402 within the IL-19 gene. The presence of five haplotypes with a frequency of ≥ 1 was estimated. The haplotypes accounted for 80% of all haplotypes in pooled samples. HT3 ACG haplotype was higher in controls as compared to psoriasis patients which shows that it may have decreased risk of psoriasis OR=0.41; (95% CI=0.20- 0.84). No other significant association was seen in remaining haplotypes.
| Haplotype | 42232 | 38386 | 35402 | Cases (%) |
Control (%) |
OR 95% CI | P |
|---|---|---|---|---|---|---|---|
| HT 1 | C | C | G | 22.13 | 25.25 | 1 | |
| HT 2 | C | T | G | 17.68 | 21.50 | 0.96 (0.60 – 1.53) | 0.88 |
| HT 3 | A | C | G | 6.80 | 18.50 | 0.41 (0.20 – 0.84) | 0.016 |
| HT 4 | C | C | C | 15.5 | 8.50 | 1.62 (0.94 – 2.78) | 0.084 |
| HT 5 | A | T | G | 12.14 | 10.50 | 1.28 (0.72 – 2.26) | 0.41 |
Statistically significant p<0.05
Table 5. Possible haplotype frequencies of IL 19 gene in cases and controls.
Disclosure
Psoriasis is regarded as an autoimmune disease in which genetic and environmental factors play a significant role. Psoriasis is characterised by plaques of red (erythematous), scaly and well-demarcated skin lesions formed due to abnormal differentiation and hyperproliferation of epidermal keratinocytes affecting 2-3% population worldwide (Shai A, Vardy D, Zvulunov A, 2002; Baliwag J, Barnes DH, Johnston A; 2015). Vascular hyperplasia and a rich immune cell penetrate in the dermis complete the histological aspects of psoriatic plaques (Wolk K, Kunz S, Asadullah K, Sabat R, 2002; Schon MP, Boehncke WH, Brocker EB, 2009; Wang H, Peters T, Kess D, Sindrilaru A, 2006). IL-19 is the pro-inflammatory cytokine that is highly elevated in psoriasis and is strongly suppressed by organization of IL-4 during the improvement of psoriasis (Ghoreschi K, Mrowietz U, Röcken M, 2003). IL-19 shares the same receptor as that of IL- 20 which shows that IL-19 may have somewhat overlapping biological activities with IL-20. In vitro data have suggested that IL-19 induces TNF-α and IL-6 production and apoptosis in monocytes (Liao YC, Liang WG, Chen FW, Hsu JH, 2002). TNF-α and IL-6 and has been clearly demonstrated in the pathogenesis of psoriasis. IL-19 and IL-20 has been detected in expression of focal suprapapillary epidermal (Takahashi H, Tsuji H, Hashimoto Y, Ishida Yamamoto A, 2010). These studies suggest that IL-19 and IL -20 may play a pathogenic role in psoriasis. In our study we found IL-19 -35402 C/C (rs2243158) genotype is associated with increased risk of psoriasis but we don’t found significant association of IL-19 -42232 C/A (rs2243188), -38386 T/C (rs2073186) with psoriasis. In contrary to our results Kingo et al. demonstrated that minor alleles of the IL-19 gene SNPs (rs2243188, rs2243169 and rs2243158) revealed protective effect to psoriasis, especially to late-onset disease and type II phenotype in estonian population (Koks S, Kingo K, Vabrit K, Rätsep R, 2005; Kingo K, Mössner R, Koks S, Rätsep R, 2007). Although study on European population reveals that no significant difference ofIL-19 gene SNPs was seen in psoriasis patients and controls (Galimova E, Khusnutdinova E Rätsep, R, Traks T, 2017). We could not compare our results with Asian population because till dateno studyhave been reported to our knowledge. HT3ACG haplotype was higher in controls as compared to psoriasis patients which shows that it may have Protective role OR=0.41; (95%CI=0.20- 0.84). Researcher found that HT7 CACCGGAA is related to increased risk (OR=2.548) for psoriasis in sample of unrelated patients and controls (Kingo K, Mössner R, Koks S, Rätsep R, 2007). Our findings are in concordance with other investigators (Ashcroft D, Li Wan Po A, Williams H, Griffiths C, 1998; Finlay A, Khan G, Luscombe D, Salek M, 1990). Psoriasis is a complex autoimmune disease with multiple genes and proteins being involved in its pathogenesis. Despite the efforts performed to understand mechanisms of psoriasis pathogenesis and to identify diagnostic and prognostic targets, disease-specific and effective biomarkers were still not available. The little understanding gained through this study on susceptibility to psoriasis may help to identify high risk individual and predictive model for preventive strategies.
Conclusion
The study provides first evidence regarding IL-19 gene and psoriasis in north Indian population. The reason for contrary results obtained from several studies remains ambiguous and might be attributed to differences in ethnic background and the selection of population studied, difference in sample size and gene-environment interaction. Research with large samples should be done in order to determine the exact role of IL-19 polymorphisms in the immunopathogenesis of psoriasis.
Acknowledgment
We thank all the technical and non-technical staff of Department of Dermatology, Venerology and Leprology, Post Graduate Institute of Medical Education and Research Chandigarh and Department of Human Genetics, Punjabi University, Patiala for their valuable cooperation and help during the course of study. The study was supported by Indian Council of Medical Research, New Delhi in the form of major research project (N. 61/13/2011-BMS).
Conflicts of interest
None.
About the Authors
Corresponding Author
Rajinder Kaur
Department of Human Genetics, Punjabi University, Patiala, Punjab, India
- Email:
- rajinderkaur@pbi.ac.in
References
- Gudjonsson J, Johnston A, Sigmundsdottir H, Valdimarsson H (2004) Immunopathogenic mechanisms in psoriasis. Clinical & Experimental Immunology 135: 1-8. https://doi.org/10.1111/j.1365-2249.2004.02310.x
- Finch CE and Crimmins EM (2004) Inflammatory exposure and historical changes in human life-spans. Science 305: 1736-1739. https://doi.org/10.1126/science.1092556
- Wani A, Ganai BA, Akhtar T, Narang T, et al. (2017) Association of proinflammatory cytokine IL-20 gene polymorphism with psoriasis in north Indian population. Egyptian Journal of Medical Human Genetics 19:201-205. https://doi.org/10.1016/j.ejmhg.2017.09.002
- Dogra S and Yadav S (2010) Psoriasis in India: Prevalence and pattern. Indian Journal of Dermatology, Venereology, and Leprology 76: 595-601. https://doi.org/10.4103/0378-6323.72443
- Gallagher G, Dickensheets H, Eskdale J, Izotova LS, et al. (2000) Cloning, expression and initial characterization of interleukin-19 (IL-19), a novel homologue of human interleukin-10 (IL-10). Genes Immun1: 442-450. https://doi.org/10.1038/sj.gene.6363714
- Wolk K, Kunz S, Asadullah K, Sabat R (2002) Cutting edge: immune cells as sources and targets of the IL-10 family members? J Immunol 168: 5397-5402. https://doi.org/10.4049/jimmunol.168.11.5397
- Gallagher G, Dickensheets H, Eskdale J, Izotova L, et al. (2000) Cloning, expression and initial characterisation of interleukin-19 (IL-19), a novel homologue of human interleukin-10 (IL-10). Genes and immunity 1: 442. https://doi.org/10.1038/sj.gene.6363714
- Koks S, Kingo K, Rätsep R, Karelson M (2004) Combined haplotype analysis of the interleukin-19 and-20 genes: relationship to plaque-type psoriasis. Genes and immunity 5: 662-667. https://doi.org/10.1038/sj.gene.6364141
- Dumoutier L, Van Roost E, Colau, D, Renauld JC (2000) Human interleukin-10-related T cell-derived inducible factor: molecular cloning and functional characterization as an hepatocyte-stimulating factor. Proceedings of the National Academy of Sciences 97: 10144-10149. https://doi.org/10.1073/pnas.170291697
- Witte E, Kokolakis G, Witte K, Philipp S Wittig (2014) IL-19 is a component of the pathogenetic IL-23/IL-17 cascade in psoriasis. Journal of Investigative Dermatology 134: 2757-2767. https://doi.org/10.1038/jid.2014.308
- Liao YC, Liang WG, Chen FW, Hsu JH (2002) IL-19 induces production of IL-6 and TNF-α and results in cell apoptosis through TNF-α. The Journal of Immunology 169: 4288-4297. https://doi.org/10.4049/jimmunol.169.8.4288
- Franceschi C, Bonafè M, Valensin S, Olivieri F (2000) Inflamm├ó┬?┬Éaging: an evolutionary perspective on immunosenescence. Annals of the New York Academy of Sciences 908: 244-254. https://doi.org/10.1111/j.1749-6632.2000.tb06651.x
- Langley RG and Ellis CN (2004) Evaluating psoriasis with psoriasis area and severity index, psoriasis global assessment, and lattice system physician's global assessment. Journal of the American Academy of Dermatology 51: 563-569. https://doi.org/10.1016/j.jaad.2004.04.012
- Sambrook J and Russell DW (2006) Purification of nucleic acids by extraction with phenol:chloroform. CSH Protoc 1: pdb.prot4455. https://doi.org/10.1101/pdb.prot4455
- Kingo K, Koks S, Nikopensius T, Silm H (2004) Polymorphisms in the interleukin-20 gene: relationships to plaque-type psoriasis. Genes and immunity 5: 117-121. https://doi.org/10.1038/sj.gene.6364046
- Wani A, Ganai BA, Akhtar T, Narang T (2018) Association of proinflammatory cytokine IL-20 gene polymorphism with psoriasis in north Indian population. Egyptian Journal of Medical Human Genetics 19: 201-205. https://doi.org/10.1016/j.ejmhg.2017.09.002
- Shai A, Vardy D, Zvulunov A (2002) Psoriasis, biblical afflictions and patients' dignity. Harefuah 141: 479-482
- Baliwag J, Barnes DH, Johnston A (2015) Cytokines in psoriasis. Cytokine 73: 342-350. https://doi.org/10.1016/j.cyto.2014.12.014
- Schon MP, Boehncke WH, Brocker EB (2009) Psoriasis: clinical manifestations, pathogenesis and therapeutic perspectives. Discovery medicine 5: 253-258.
- Wang H, Peters T, Kess D, Sindrilaru A (2006) Activated macrophages are essential in a murine model for T cell–mediated chronic psoriasiform skin inflammation. The Journal of clinical investigation 116: 2105-2114. https://doi.org/10.1172/jci27180
- Ghoreschi K, Mrowietz U, Röcken M (2003) A molecule solves psoriasis? Systemic therapies for psoriasis inducing interleukin 4 and Th2 responses. Journal of molecular medicine 81: 471-480. https://doi.org/10.1007/s00109-003-0460-9
- Takahashi H, Tsuji H, Hashimoto Y, Ishida Yamamoto A (2010) Serum cytokines and growth factor levels in Japanese patients with psoriasis. Clinical and experimental dermatology 35: 645-649. https://doi.org/10.1111/j.1365-2230.2009.03704.x
- Koks S, Kingo K, Vabrit K, Rätsep R (2005) Possible relations between the polymorphisms of the cytokines IL-19, IL-20 and IL-24 and plaque-type psoriasis. Genes and immunity 6: 407-415. https://doi.org/10.1038/sj.gene.6364216
- Kingo K, Mössner R, Koks S, Rätsep R (2007) Association analysis of IL19, IL20 and IL24 genes in palmoplantar pustulosis. British Journal of Dermatology 156: 646-652. https://doi.org/10.1111/j.1365-2133.2006.07731.x
- Galimova E, Khusnutdinova E Rätsep, R, Traks T (2017) Interleukin├ó┬?┬É10 family cytokines pathway: genetic variants and psoriasis. British Journal of Dermatology 176: 1577-1587. https://doi.org/10.1111/bjd.15363
- Ashcroft D, Li Wan Po A, Williams H, Griffiths C (1998) Quality of life measures in psoriasis: a critical appraisal of their quality. Journal of clinical pharmacy and therapeutics 23: 391-398. https://doi.org/10.1046/j.1365-2710.1998.00181.x
- Finlay A, Khan G, Luscombe D, Salek M (1990) Validation of sickness impact profile and psoriasis disability index in psoriasis. British Journal of Dermatology 123: 751-756. https://doi.org/10.1111/j.1365-2133.1990.tb04192.x
Keywords:
Download:
Full PDF- Share This