Preliminary analysis on the developmental transcriptomes of swallowtail butterfly Papilio polytes (Lepidoptera: Papilioidae)
Received: February 11, 2018
Accepted: April 06, 2018
Published: April 12, 2018
Genet.Mol.Res. 17(2): gmr16039902
DOI: 10.4238/gmr16039902
Abstract
The butterflies are one of the insect groups which undergo typical complete metamorphosis in their life history. However, up to now, little is known about the genomic mechanism that regulates the butterfly metamorphosis. In this study, using a swallowtail butterfly P. polytes as a model organism, a high-throughput sequencing platform was employed to perform the transcriptome and gene expression analyses in order to explore the P. polytes transcriptome features during different developmental stages. The results showed that approximately 398 million useful (Q20) reads were assembled into groups of 14698 (L1), 14264 (L2), 15084 (L3), 15520 (P1), 15052 (P2), 15720 (P3) and 15709 (A1), 14668 (A2), 16152 (A3) genes, respectively, with 58.19% to 67.11% of the data successfully mapped to the reference genome; the transcriptome change analysis via the DEGs Package revealed that dramatic gene expression differences were presented among the different developmental stages, that is, totally, 1162, 891 and 1723 genes were differentially expressed between adult and pupal, adult and larval, larval and pupal stages, respectively, with a number of these differentially expressed genes associated with the functions of digestion, cuticularization, chemoreception, wing formation, and so on. These differentially expressed genes and potential candidate genes required for butterfly metamorphosis by comparative transcriptomics may shed some new insights on molecular mechanisms underlying complete metamorphosis.
Introduction
Metamorphosis as a life-history strategies of insects, is an attractive and highly successful biological adaptation (Truman and Riddiford, 1999). The insects of complete metamorphosis such as butterflies, moths, flies and bees, undergo a series of continuous changes through their lifetimes, that is, in descending order they go through egg to larval, pupal, and adult stages in their different developmental periods (Resh and Cardé, 2009). The typical pupa which spans the larval and adult periods, is undergoing a series of intensely active changes to the last formation of various adult tissues and organs, including two pairs of wings (Xu, 2009), although the surface seems statically unchanged.
Insects are the most specious and evolutionarily successful groups in animals on earth, this is caused by the interaction of various factors, and among which the complex changeable metamorphosis is undoubtedly an important one (Malmstrom, 2012). In paleontological view, the occurrence and development of complete metamorphosis on the earth is probably related to the radically climatic transition between the Carboniferous and Permian. Changes in environmental factors can alter the activity of the endocrine system of organisms which can influence a series of physiological and biochemical changes, as well as morphological and behavioral alterations of animals. Thus the holometabolism (pupae formation from larvae to adults) may have developed rapidly due to the strong resistance of pupae to various environmental changes, especially, the rapid and violent climate change (Jia and Zhang, 1999).
In recent years, next-generation high-throughput DNA sequencing techniques have provided fascinating opportunities in the life sciences and dramatically improved the efficiency and cost-effectiveness of gene discovery (Ansorge, 2009), and for that reason these techniques has been widely used in the study of diverse organisms such as human beings (Lappalainen et al., 2013), plants (Garg and Jain, 2013; Ichihashi et al., 2015), agricultural pests ( Zhang et al., 2014; Zhang et al., 2016) to clarify the dynamic changes of gene expression and regulation during different developmental staged of these model and non-model organisms (Filichkin et al., 2010; Oppenheim et al., 2015). Up to the present, dozens of lepidopteran transcriptomes have been sequenced, however, only a few of developmental transcriptomes are available (Chen et al., 2010; Chen et al., 2012; Li et al., 2013; Qi et al., 2016; Zheng et al., 2012). Li et al. (2013) compared the transcriptomes of four developmental stages of the night moth Athetis lepigone and found that the most dramatic differences in gene expression were detected in the transitions from one stage to another stage. Some of these differentially expressed genes are related to cuticle and wing formation as well as the growth and development, and these expression differences were consistent with characteristic features of each life stage, for example, cuticular protein genes were up-regulated in larvae and down-regulated in pupae. Qi et al. (2016) sequenced the transcriptomes of different development stages of the small white butterfly Pieris rapae, and the results showed that a total of 21,883 differentially expressed unigenes were detected across the developmental stages, most of which were found between the egg and first larval stages, and total 32 heat shock protein (Hsp) genes with similar expression pattern were identified. In Hymenoptera, Chen et al. (2012) contrasted the transcriptomes of full-sister queen (QL) and worker-destined (WL) larvae of the honeybee using high-throughput RNA-Seq, and found that more than 4,500 genes with different expressions were existed between the two types larvae, of which more than 70% were upregulated in tha QL larvae.
The Common Mormon, Papilio polytes (Lepidoptera: Papilioidae) is a member of swallowtail butterfly species widely distributed in Asia. They undergo a series of striking changes through its lifetime as they mature from egg to adult (Chou, 1994). P. polytes as a kind of typical complete metamorphosis insects, the life of P. polytes experienced a very complex biological process, but little is known about the mechanism that regulates the development of P. polytes. Additionally, functional transcriptome information may lead to profound insights of the biology of P. polytes. In this study, taken P. polytes as a model organism, we generated a substantial dataset of transcript reads at their different developmental stages from larvae to adults, by using Illumina NextSeq500 RNA-Seq technique, in order to reveal the gene expression characteristics and clarify the functions of related genes or gene families in their different developmental stages.
Materials and Methods
Materials
The larval, pupal and adult individuals of P. polytes were collected at the campus of Anhui Normal University (Wuhu, China) in May and June of 2017. Test samples used in this study were: (1) a tissue mixture of third instar larvae; (2) a tissue mixture of pupae and (3) a tissue mixture of adults. In this study, adult male P. polytes were used for this study. Each sample after collection was immediately frozen in liquid nitrogen and stored at -80oC until RNA extraction. Three biological repeats for each stage: larvae (L1, L2, L3), pupae (P1, P2, P3) and adults (A1, A2, A3) were set in the experiments to exclude the artificial errors.
RNA isolation, cDNA library preparation and illumina sequencing
Total RNA from tissues of larvae, pupae and adults was extracted separately according to the manufacturer’s protocol. RNA integrity was confirmed by the Agilent 2100 Bioanalyzer (Agilent Technologies), and cDNA library construction and Illumina sequencing were subsequently performed at Personal Biotechnology Co., Ltd., Shanghai China. Briefly, Magnetic beads with oligo-dT were used to combine the poly-A of the mRNA for purifying the mRNA from the total RNA. The mRNA was then mixed with fragmentation buffer to obtain 200-300 bp short fragments. The fragments were used to synthesize first-strand cDNA with random primers, and first-strand cDNA was transformed into double-strand cDNA by using RNase H and DNA polymerase I. When the second chain cDNA began to syntheze, the bases of these T should be replaced with U, so as to achieve the aim of chain specific library. Library fragments were purified with a QIAquick PCR Extraction Kit (Qiagen, Valencia, CA, USA), and the sequencing library was constructed using polymerase chain reaction (PCR). The multiplex cDNA libraries were checked using PicoGreen (Quantifluor™-ST fluorometer E6090, Promega, CA, USA) and fluoro spectrophotometry (Quant-iT PicoGreen dsDNA Assay Kit; Invitrogen, P7589) and quantified with Agilent 2100 (Agilent 2100 Bioanalyzer, Agilent, 2100; Agilent High Sensitivity DNA Kit, Agilent, 5067–4626). The synthesized cDNA libraries were then normalized to a 10 nM, and the sequencing library was gradually diluted and quantified to 4–5 pM and sequenced using the Illumina NextSeq™ 500 platform instrument with paired-end (PE) sequencing method.
Gene expression analysis
In order to minimize sequencing mistakes, adaptor sequences and low reads were filtered out prior to mapping, the reference genome index was established by Bowtie2 (Langmead et al., 2009), and then the filtered reads were compared to the reference genome using Tophat2 (Kim et al., 2013) (http://tophat.cbcb.umd.edu/). The “RPKM” method (reads per kilo bases per million reads) was used for normalization with the total mapped reads count and the gene length to quantitate the level of gene expression, and results less than 0.005 were excluded (Mortazavi et al., 2008). The significance was determined by normalizing the raw reads and calculating the P-value using DESeq (Anders, 2012). Genes with fold change>2 or <0.5 and P-value<0.05 were identified as differentially expressed gene (DEG) (Anders and Huber, 2010).
The Blast2GO (Conesa et al., 2005) program were used to compare genes with GO (Gene Ontology) database (http://www.geneontology.org/), and the GO terms for each gene were submitted to Web Gene Ontology Annotation Plot (WEGO) for further classification (Ashburner et al., 2000). The genes significantly different from the corresponding library were searched against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database to determine the pathways (Kanehisa et al., 2004).
Results and Discussion
Raw data processing
In total, 404 million reads were generated from the nine libraries, and 398 million useful (Q20) reads were selected for further analysis. The useful reads were mappted to the P. polytes genome (Nishikawa et al., 2015) using tophat software and the results showed that 58.19% to 67.11% of reads were successfully mapped. Among them, 69.91% to 87.67% could be mapped to unigenes, 95.13% to 96.59% were successfully mapped to exonic regions (Table 1). Totally, 16360 genes were identified, and among these genes, 14698, 14264, 15084, 15520, 15052, 15720, 15709, 14668 and 16152 genes were identified in the L1, L2, L3, P1, P2, P3, A1, A2, A3 libraries, respectively (Table 1).
| Sample | Raw Reads | Useful Reads | Total Mapped (%) | Mapped to Exon (%) | Mapped to Gene% | Number of genes | Number of novel genes |
|---|---|---|---|---|---|---|---|
| L1 | 42406996 | 41728282 | 62.11 | 96.39 | 88.29 | 14698 | 4207 |
| L2 | 42016536 | 41451978 | 65.36 | 96.18 | 84.63 | 14264 | 3909 |
| L3 | 44720010 | 44071764 | 64.92 | 96.59 | 85.59 | 15084 | 4295 |
| P1 | 48175076 | 47464216 | 63.64 | 96.21 | 87.67 | 15520 | 5133 |
| P2 | 45990738 | 45322112 | 61.39 | 96.07 | 85.63 | 15052 | 4783 |
| P3 | 44468890 | 43851132 | 63.06 | 95.95 | 87.11 | 15720 | 5346 |
| A1 | 46981392 | 46309912 | 63.13 | 95.28 | 84.52 | 15709 | 5295 |
| A2 | 45032864 | 44373054 | 67.11 | 95.13 | 84.96 | 14668 | 4515 |
| A3 | 44206110 | 43561518 | 58.19 | 92.66 | 69.91 | 16152 | 5384 |
Table 1. Summary statistics of clean reads in the P. polytes transcriptomes.
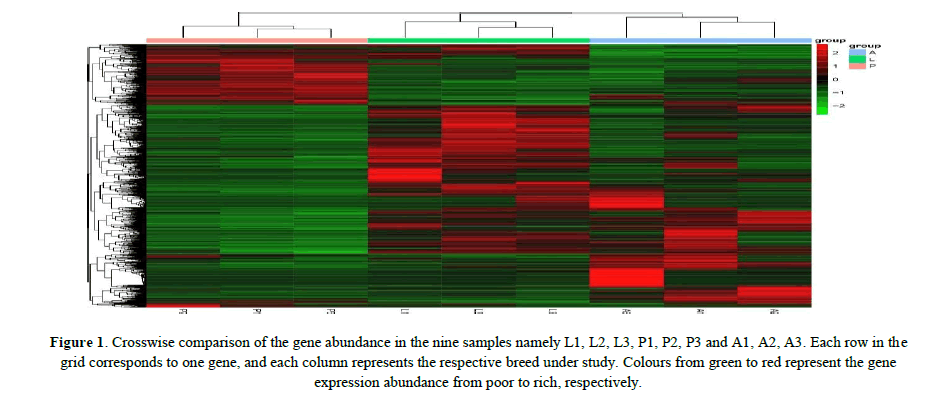
The DEGs of three stages were analyzed through heat maps, and hierarchical clusters were generated to obtain a global view of the DEGs which were illustrated using a grid of colored tiles. A crosswise comparison of 9 samples were performed through cluster analysis, and the results showed that the samples from the same stage were clustered into the corresponding group; the genes detected in the different groups were clearly separated; the increase or decrease in transcript abundance in the pupae and adults was significantly different from those in the larvae; in addition, P1, P2 and P3 were basically the same, L1, L2 and L3 were similar in general, while A1, A2 and A3 were somewhat different with each other. These results indicated that the gene expressions changed dramatically during different developmental stages from larvae to adults, and overally larvae and aldults harbored higher gene expression abundances than those in the pupae (Figure 1).
Figure 1: Crosswise comparison of the gene abundance in the nine samples namely L1, L2, L3, P1, P2, P3 and A1, A2, A3. Each row in the grid corresponds to one gene, and each column represents the respective breed under study. Colours from green to red represent the gene expression abundance from poor to rich, respectively.
Differentially expressed genes (DEGs) during P. polytes development
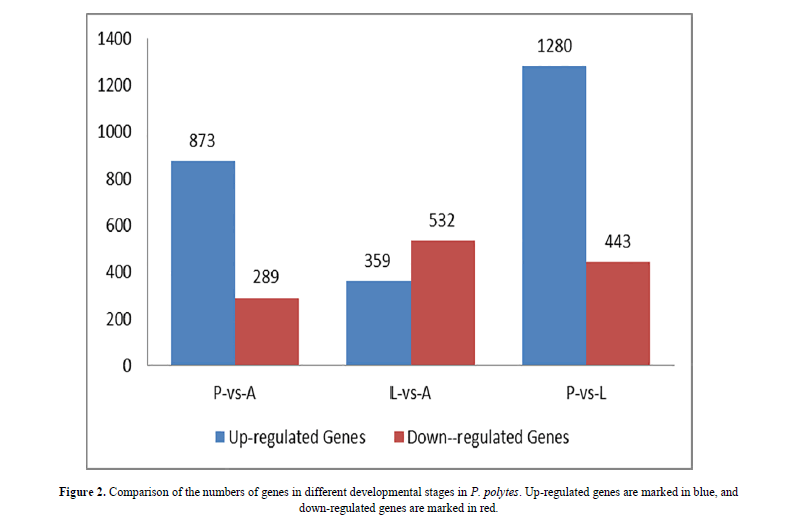
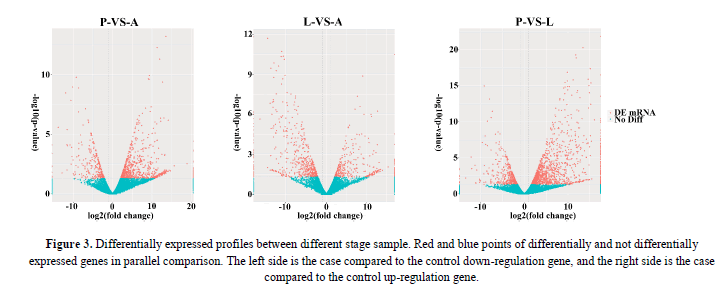
Differences in gene expression and DEGs were examined and identified at three stages during P. polytes development by pairwise comparisons of the three libraries. The numbers of up- and down-regulated genes were also calculated between each pair of life-stages. The statistical results were shown in Figure 2 and Figure 3, with the top 10 up and down regulated genes listed in Table 2. Apparently, over 70% of the DEGs were more highly expressed in the larvae (L) and adults (A) than in the pupae (P).
Figure 3: Differentially expressed profiles between different stage sample. Red and blue points of differentially and not differentially expressed genes in parallel comparison. The left side is the case compared to the control down-regulation gene, and the right side is the case compared to the control up-regulation gene.
By comparison between the larval and pupal transcriptomes, 1,723 significantly differentially expressed genes were detected, including 1280 (74.3%) up-regulated and 443 (25.7%) down-regulated genes in the larval library. Seven of the top ten up-regulated genes in larval transcriptomes were matched with the orthologs of P. polytes genome: one uncharacterized protein LOC106104536, six with Predicted functions (bile salt-activated lipase-like, trypsin, alkaline C-like, membrane alanyl aminopeptidase-like, fibroin light chain-like, aminopeptidase N-like and ejaculatory bulb-specific protein 3-like precursor); the other two were matched with the sodium-dependent nutrient amino acid transporter 1 and uncharacterized protein LOC106119902 in Papilio xuthus respectively; the last one was matched with of the membrane alanyl aminopeptidase in Papilio machaon. Besides, nine of the top ten down-regulated genes in larval transcriptomes (mushroom body large-type Kenyon cell-specific protein 1 isoform X2, delta-like protein 1 isoform X1, putative cyclin-dependent serine/threonine-protein kinase DDB_G0272797/DDB_G0274007 isoform X1, uncharacterized protein LOC106100118, LOC106100129, LOC106099981, LOC106108048, LOC106109383 and LOC106108920) were matched with P. polytes, one was matched with the hypothetical protein RR46_06374 of P. xuthus (Table 2).
| GeneID | P | L | p-value | Description |
|---|---|---|---|---|
| Up | ||||
| P. polytes Gene0001048 |
0 | 8243.783 | 1.34336E-22 | Sodium-dependent nutrient amino acid transporter 1 [Papilio xuthus] |
| P. polytes Gene0003613 |
0 | 1166.543 | 2.98573E-17 | Predicted: uncharacterized protein LOC106104536 [Papilio polytes] |
| P. polytes Gene0010655 |
0 | 1410.063 | 2.71489E-14 | Predicted: bile salt-activated lipase-like [Papilio polytes] |
| P. polytes Gene0002153 |
1.442 | 56780.33 | 3.77289E-16 | Membrane alanyl aminopeptidase [Papilio machaon] |
| P. polytes Gene0008568 |
0.241 | 9246.798 | 3.57096E-13 | Predicted: trypsin, alkaline C-like [Papilio polytes] |
| P. polytes Gene0002152 |
0.721 | 25739.1 | 4.10209E-18 | Predicted: membrane alanyl aminopeptidase-like [Papilio polytes] |
| P. polytes Gene0002149 |
0.722 | 22323.06 | 8.7855E-15 | Predicted: aminopeptidase N-like [Papilio polytes] |
| P. polytes Gene0005536 |
0.276 | 7859.919 | 9.23473E-16 | ejaculatory bulb-specific protein 3-like precursor [Papilio polytes] gi|389610705|dbj|BAM18964.1| ejaculatory bulb protein III [Papilio polytes] |
| P. polytes Gene0010477 |
1.444 | 36878.47 | 1.20068E-16 | Predicted: fibroin light chain-like [Papilio polytes] |
| P. polytes Gene0006740 |
0.240 | 2646.283 | 4.88662E-21 | Predicted: uncharacterized protein LOC106119902 [Papilio xuthus] |
| Down | ||||
| P. polytes Gene0007402 |
17369.33 | 3.625 | 7.65547E-06 | Predicted: uncharacterized protein LOC106100118 [Papilio polytes] |
| P. polytes Gene0007403 |
5012.981 | 6.163 | 7.20085E-08 | Predicted: uncharacterized protein LOC106100129 [Papilio polytes] |
| P. polytes Gene0002077 |
3713.067 | 6.256 | 1.02074E-15 | Predicted: mushroom body large-type Kenyon cell-specific protein 1 isoform X2 [Papilio polytes] |
| P. polytes Gene0007573 |
1784.42 | 3.443 | 8.82427E-09 | Predicted: uncharacterized protein LOC106099981 [Papilio polytes] |
| P. polytes Gene0006285 |
1362.624 | 2.839 | 1.34905E-07 | Predicted: delta-like protein 1 isoform X1 [Papilio polytes] |
| P. polytes Gene0009792 |
19061.27 | 52.460 | 8.05246E-11 | Predicted: putative cyclin-dependent serine/threonine-protein kinase DDB_G0272797/DDB_G0274007 isoform X1 [Papilio polytes] |
| P. polytes Gene0000508 |
40599.11 | 112.406 | 1.95646E-07 | Predicted: uncharacterized protein LOC106108048 [Papilio polytes] |
| P. polytes Gene0005657 |
3210.683 | 12.218 | 6.79535E-14 | hypothetical protein RR46_06374 [Papilio xuthus] |
| P. polytes Gene0011456 |
3931.157 | 19.508 | 8.18186E-07 | Predicted: uncharacterized protein LOC106109383 [Papilio polytes] |
| P. polytes Gene0011800 |
9426.428 | 47.383 | 2.54548E-11 | Predicted: uncharacterized protein LOC106108920 [Papilio polytes] |
| L | A | Description | ||
| Up | ||||
| P. polytes Gene0000001 |
0 | 1171.562 | 3.2993E-11 | female-specific doublesex isoform F1 [Antheraea mylitta] |
| P. polytes Gene0004183 |
1.025398 | 6976.514 | 5.92951E-07 | Predicted: UDP-glucuronosyltransferase 2C1-like [Papilio polytes] |
| P. polytes Gene0006367 |
1.028 | 2493.631 | 1.48393E-06 | hypothetical protein RR48_14296 [Papilio machaon] |
| P. polytes Gene0006347 |
2.572 | 5806.303 | 1.01609E-06 | Predicted: uncharacterized protein LOC106101520 [Papilio polytes] |
| P. polytes Gene0006589 |
0.734 | 494.2101 | 1.02014E-06 | Predicted: nose resistant to fluoxetine protein 6-like [Papilio polytes] |
| P. polytes Gene0002077 |
4.838 | 2612.353 | 1.38538E-09 | Predicted: mushroom body large-type Kenyon cell-specific protein 1 isoform X2[Papilio polytes] |
| P. polytes Gene0006369 |
16.110 | 5514.288 | 2.58682E-07 | Predicted: uncharacterized protein LOC106101491 [Papilio polytes] |
| P. polytesGene0001904 | 13.507 | 2933.525 | 4.52601E-08 | Predicted: inducible metalloproteinase inhibitor protein-like [Papilio polytes] |
| P. polytes Gene0009269 |
7.727 | 1137.037 | 4.21618E-07 | Predicted: protein msta, isoform A [Papilio polytes] |
| P. polytes Gene0011372 |
0.368 | 3325.782 | 0.004127324 | Predicted: troponin C, isoform 1 [Papilio polytes] |
| Down | ||||
| P. polytesGene0007045 | 1681.93 | 0 | 1.55411E-12 | Predicted: 4-coumarate--CoA ligase 1-like [Papilio polytes] |
| P. polytesGene0008568 | 7121.047 | 0.316 | 2.03899E-12 | Predicted: trypsin, alkaline C-like [Papilio polytes] |
| P. polytesGene0008454 | 4114.453 | 0.316 | 3.59609E-10 | Predicted: gastrolith matrix protein-like [Papilio polytes] |
| P. polytesGene0010407 | 55872.09 | 6.579 | 8.0936E-08 | Predicted: carboxypeptidase B-like [Papilio polytes] |
| P. polytesGene0009084 | 53137.33 | 6.505 | 1.13051E-07 | Predicted: uncharacterized protein LOC106111207 [Papilio polytes] |
| P. polytesGene0003648 | 37073.76 | 4.610 | 1.73304E-09 | Predicted: flexible cuticle protein 12-like [Papilio polytes] cuticular protein PpolCPR4B [Papilio polytes] |
| P. polytesGene0002670 | 20328.58 | 2.620 | 1.42466E-10 | Predicted: uncharacterized protein LOC106105319 [Papilio polytes] |
| P. polytesGene0000067 | 22453.83 | 4.306 | 5.43216E-10 | Predicted: glycine-rich protein 1-like [Papilio polytes] |
| P. polytesGene0006748 | 14831.24 | 2.856 | 2.95024E-08 | bilin-binding protein-like precursor [Papilio polytes] bilin binding protein 1 [Papilio polytes] |
| P. polytesGene0010655 | 1085.122 | 0.309707 | 2.47302E-10 | Predicted: bile salt-activated lipase-like [Papilio polytes] |
| P | A | Description | ||
| Up | ||||
| P. polytesGene0001899 | 0 | 1200.988 | 3.76989E-05 | hypothetical protein RR48_11198 [Papilio machaon] |
| P. polytesGene0005152 | 0 | 1755.63 | 0.000216058 | Predicted: synaptic vesicle glycoprotein 2A-like [Papilio polytes] |
| P. polytesGene0001552 | 0 | 1513.632 | 0.00708644 | Predicted: retinoid-inducible serine carboxypeptidase-like [Papilio polytes] |
| P. polytesGene0002988 | 0 | 5086.073 | 0.008164307 | hypothetical protein RR46_02520 [Papilio xuthus] |
| P. polytesGene0005469 | 0 | 4831.032 | 0.009456105 | Predicted: carboxypeptidase A1-like [Papilio polytes] |
| P. polytesGene0011731 | 0.243 | 120187.8 | 0.002859641 | Predicted: uncharacterized protein LOC106709423 [Papilio machaon] |
| P. polytesGene0003329 | 0.243 | 12582.03 | 0.004340028 | Predicted: pancreatic triacylglycerol lipase-like [Papilio polytes] |
| P. polytesGene0010641 | 0.490 | 8866.998 | 6.49639E-07 | Predicted: serine proteases 1/2-like [Papilio polytes] |
| P. polytesGene0002750 | 0.243 | 2870.321 | 5.96095E-14 | Predicted: uncharacterized protein LOC106105328 [Papilio polytes] |
| P. polytesGene0011796 | 0.243 | 2454.32 | 0.000516075 | Predicted: ammonium transporter Rh type B-B [Papilio polytes] |
| Down | ||||
| P. polytesGene0000070 | 545.402 | 0 | 0.00007581 | Predicted: protein doublesex isoform X2 [Papilio xuthus] |
| P. polytesGene0001023 | 283.627 | 0 | 0.00039516 | Predicted: D-beta-hydroxybutyrate dehydrogenase, mitochondrial [Papilio polytes] |
| P. polytesGene0007402 | 17761.604 | 1.265 | 2.5637E-06 | Predicted: uncharacterized protein LOC106100118 [Papilio polytes] |
| P. polytesGene0007403 | 5116.827 | 1.335 | 3.3E-09 | Predicted: uncharacterized protein LOC106100129 [Papilio polytes] |
| P. polytesGene0000399 | 11501.824 | 3.793 | 0.000070974 | Predicted: uncharacterized protein LOC106107989 [Papilio polytes] |
| P. polytesGene0009068 | 20058.079 | 6.820 | 4.0011E-06 | Predicted: uncharacterized protein LOC106111008 [Papilio polytes] |
| P. polytesGene0002954 | 14830.720 | 5.200 | 8.90061E-05 | Predicted: basic juvenile hormone-suppressible protein 2-like [Papilio polytes] |
| P. polytesGene0000508 | 41392.585 | 27.065 | 1.41E-08 | Predicted: uncharacterized protein LOC106108048 [Papilio polytes] |
| P. polytesGene0006861 | 12410.295 | 9.280 | 0.000122486 | Predicted: actin cytoskeleton-regulatory complex protein PAN1-like [Papilio polytes] |
| P. polytesGene0004084 | 42035.774 | 33.528 | 0.000721261 | Predicted: basic juvenile hormone-suppressible protein 1-like [Papilio polytes] |
(Note: In L-vs-P, Dark gray represents the gene that is up-regulated in larvae, and the light gray represents the gene that is up-regulated in pupae. In L-vs-A, The light blue represents up-regulated genes in adults, and the dark blue represents increased in larvae. In P-vs-A, Dark red represents the gene that is up-regulated in adults, and the light red represents up-regulated gene in pupae).
Table 1. The top 10 up or down-regulated genes identified among Pupae (P) vs. larvae (L), larvae (L) vs. adults (A), and pupae (P) vs. adults (A).
By comparison between the adult and pupal transcriptomes, 1,162 genes with significantly differential expression levels were detected in adults, and among them, 873 (75.1%) up-regulated genes and 289 (24.9%) down-regulated genes were found. Seven of the top ten up-regulated genes were matched with the synaptic vesicle glycoprotein 2A-like, retinoid-inducible serine carboxypeptidase-like, carboxypeptidase A1-like, pancreatic triacylglycerol lipase-like, serine proteases 1/2-like, uncharacterized protein LOC106105328 and ammonium transporter Rh type B-B in P. polytes; three genes were matched with the hypothetical protein RR46_02520 in P. xuthus, hypothetical protein RR48_11198 and uncharacterized protein LOC106709423 in P. machaon respectively. The top ten down-regulated genes in the adult library included the following Predicted functional genes: protein doublesex isoform X2 in P. xuthus; D-beta-hydroxybutyrate dehydrogenase, mitochondrial; basic juvenile hormone-suppressible protein 2-like, basic juvenile hormone-suppressible protein 1-like and actin cytoskeleton-regulatory complex protein PAN1-like, and four uncharacterized protein LOC106100118, LOC106100129, LOC106107989, LOC106111008, LOC106108048 in P. polytes (Table 2).
A total of 891 genes (including 359 (40.3%) up-regulated genes and 532 (59.7%) down-regulated genes) were detected between larval and adult transcriptomes. Among the top ten up-regulated gene in adults, eight genes were matched with P. polytes, UDP-glucuronosyltransferase 2C1-like, uncharacterized protein LOC106101520, LOC106101491, nose resistant to fluoxetine protein 6-like, mushroom body large-type Kenyon cell-specific protein 1 isoform X2, inducible metalloproteinase inhibitor protein-like, protein msta, isoform A and troponin C isoform 1 respectively; the other two up-regulated genes were matched with the female-specific doublesex isoform F1 in silkworm Antheraea mylitta and hypothetical protein RR48_14296 in P. machaon. Among the top ten down-regulated genes in the adult transcriptome, five genes were the 4-coumarate-CoA ligase 1-like, trypsin, alkaline C-like, gastrolith matrix protein-like, carboxypeptidase B-like, flexible cuticle protein 12-like, glycine-rich protein 1-like, bile salt-activated lipase-like in P. polytes; the other two genes with unknown functions were the uncharacterized proteins LOC106111207 and LOC106105319 in P. polytes; the last one was matched with the bilin-binding protein-like precursor/bilin binding protein 1 in P. polytes (Table 2).
Functional classification of DEGs during P. polytes development
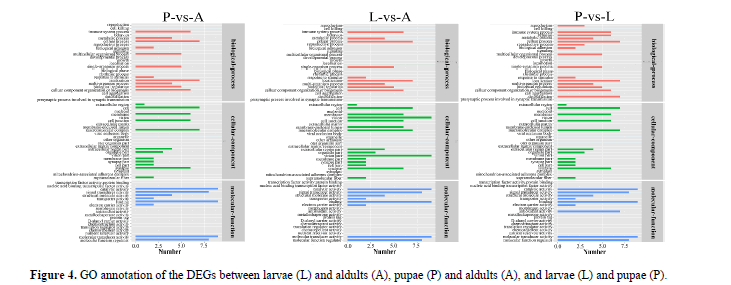
WEGO annotation of DEGs: We used GO assignments to classify the functions of DEGs in pairwise comparisons of cDNA libraries between different P. polytes developmental stages. These genes were divided into three categories: molecular function (MF), cellular component (CC) and biological process (BP). In total, 67 categories were subdivided from the primary categories: 24 categories for biological process, 24 categories for cellular component and 19 categories for molecular function. In the three GO categories, the highest proportion of annotations are cellular process and localization in the category of biological process, cell and macromolecular complex in the cellular component, catalytic activity and binding in the molecular function (Figure 4). The results showed that these significantly enriched biological functions played a key role during the growth and development of the butterflies.
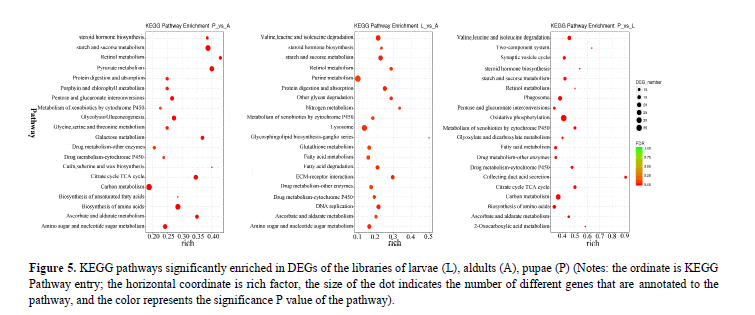
Pathway annotation of DEGs between developments: To understand the functions of the DEGs, we mapped them using the KEGG database for signaling pathways analysis. The top 20 most abundant differentially expressed signaling pathways from each compared group were listed in Figure 5. The results showed that in all the three groups, the cluster for metabolic pathways was the largest group, within which both of the fatty acids and cytochrome P450 (CYPs) were significantly enriched. Fatty acids (an important source of fuel for muscular contraction and general metabolism) and CYPs (the major enzymes involved in drug metabolism and participate in the synthesis of steroid hormones) are all closely related to the protein translation, processing modification, hormone synthesis and other aspects of metabolism in the development of P. polytes.
Figure 5: KEGG pathways significantly enriched in DEGs of the libraries of larvae (L), aldults (A), pupae (P) (Notes: the ordinate is KEGG Pathway entry; the horizontal coordinate is rich factor, the size of the dot indicates the number of different genes that are annotated to the pathway, and the color represents the significance P value of the pathway).
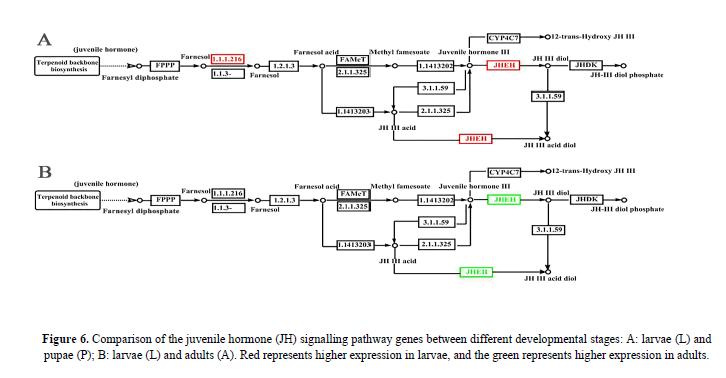
DEGs in juvenile hormone pathway
Juvenile hormone (JH) and ecdysterone are the decisive endogenous factors that induce the development of insects (Dubrovsky, 2005; Jindra & Riddiford, 2013; Truman & Riddiford, 1999, 2002). JH prevents the metamorphosis induced by the ecdysone, keeping the larvae in the larval condition (Truman and Riddiford, 2007). In the P. polytes JH pathway of this study, two enzymes were detected to have a strong effect on the JH titre, one is JHAMT (juvenile hormone acid methyltransferase) that plays a key role in regulates the last step of JH biosynthesis (Wijesekera & Dauwalder, 2016), the another is JHEH (juvenile hormone epoxide hydrolase) that is involved in primary degradation of JH. Out study showed that the JHEH gene was obviously up-regulated in larvae (L) compared with papue (P), resulting in a relatively lower JH titre in larvae (plot A, Figure 6); the JHAMT gene was not shown to be up-regulated, whereas the JHEH shown to be down-regulated in adults (A) compared with larvae (L), resulting in a higher JH titre in adults (plot B, Figure 6). These results were congruent with those of previous studies in other insect groups (Yang et al., 2011; Wijesekera and Dauwalder, 2016).
Conclusion
The P. polytes exhibits the typical developmental characteristics of holometabolous metamorphosis of insects. Our study showed that during the transitions from one to another developmental stages, the gene expressions were significantly correlated with the developmental characteristics of P. polytes. During the transition from larvae to puape, the most up-regulated genes such as membrane alanyl aminopeptidase (aminopeptidase N, APN) and ejaculatory bulb-specific protein 3-like precursor, are closely related to immune, digestive, and chemoreceptive functions respectively. For example, APNs expressed in the midgut of the insect larvae and primarily involved in dietary protein digestion (Wang and Zhang, 2005) and as receptors in Cry toxin-induced pathogenesis in insects (Bravo and Soberón, 2007); the putative cyclin-dependent serine/threonine-protein kinase was highly expressed in the pupae than in the larvae, it is a key factor in the control of cell division by modulating transcription in response to several extra- and intracellular activities in this radical transitional process (Malumbres, 2014). During the transition from puape to adults, the genes responsible for digestion, defense response, development and reproduction, such as the retinol-binding protein gene and serine proteases 1/2-like were remarkably up-regulated in the adults. For example, serine proteases are the principal digestive enzymes which provide nutrients needed for survival and fecundity (Bao et al., 2014; Zou et al., 2006), however, some genes responsible for juvenile-state keeping and metamorphosis preventing, such as the juvenile hormone-suppressible protein 2-like/1-like were highly expressed in the pupal stage, which was consistent with the results of previous studies in hemimetabolous locust Schistocerca gregaria by juvenile hormone treatment (Novak, 1969; Micciarelli, 1977; Riddiford, 2012). When larvae and adults were compared, the nose resistant to fluoxetine and the troponin C (TpnC) genes which regulates the muscle movements and carries out the flight functions were highly expressed at the adult stage (Bullard and Pastore, 2011; Herranz and Marco, 2005; Kržič et al., 2010).
In general, our study constructed nine transcriptome libraries and generated new genetic resources for the investigation of P. polytes development. This latest molecular research as well as many other in-depth studies about insect metamorphosis will shed some new lights on the underlying genomic mechanisms (Liu et al., 2017) and related evironments (Helm et al., 2017) about these complicated biological processes.
Acknowledgments
This work was supported by the Special Funds for Cultivation of Preponderant Disciplines of Anhui Normal University and funds from the State Key Laboratory of Palaeobiology and Stratigraphy (Nanjing Institute of Geology and Palaeontology, CAS) (Grant No. Y626040108).
About the Authors
Corresponding Author
Jia-sheng Hao
College of Life Sciences, Anhui Normal University, Wuhu, China 241000, China
- Email:
- jshaonigpas@sina.com
References
- Anders S (2012) Analysing RNA-Seq data with the DESeq package. Mol. Biol. 43: 1-17.
- Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biology. 11 (10): R106. https://doi.org/10.1038/npre.2010.4282.1
- Ansorge WJ (2009) Next-generation DNA sequencing techniques. New Biotechnology. 25: 195-203. https://doi.org/10.4172/2469-9853.s1-005
- AshburnerAshburner M, Ball CA, Blake JA, Botstein D, et al. (2000) Gene ontology: tool for the unification of biology. Nat Genet. 25: 25-29.
- Bao YY, Qin X, Yu B, Chen LB, et al. (2014) Genomic insights into the serine protease gene family and expression profile analysis in the planthopper, Nilaparvata lugens. BMC Genomics.15 (1): 507. https://doi.org/10.1186/1471-2164-15-507
- BravoBravo A, Gill SS, Soberón M (2007) Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon. 49 (4): 423. https://doi.org/10.1016/j.toxicon.2006.11.022
- Bullard B, Pastore A (2011) Regulating the contraction of insect flight muscle. J Muscle Res Cell Motil. 32 (5): 303-313. https://doi.org/10.1007/s10974-011-9278-1
- Chen S, Yang P, Jiang F, Wei Y, et al. (2010) De novo analysis of transcriptome dynamics in the migratory locust during the development of phase traits. PLoS One. 5 (12): e15633. https://doi.org/10.1371/journal.pone.0015633.
- Chen X, Hu Y, Zheng H, Cao L, et al. (2012) Transcriptome comparison between honey bee queen- and worker-destined larvae. Insect Biochem. Mol. Biol. 42 (9): 665-673. https://doi.org/10.1016/j.ibmb.2012.05.004
- Chou I (1994) Monographia Rhopalocerorum Sinensium. Henan Scientific and Technological Publishing House, Zhengzhou.
- Conesa A, Götz S, Garcíagómez JM, Terol J, et al. (2005) Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 21 (18): 3674-3676. https://doi.org/10.1093/bioinformatics/bti610
- Dubrovsky EB (2005) Hormonal cross talk in insect development. Trends Endocrinol Metab. 16 (1): 6-11. https://doi.org/10.1016/j.tem.2004.11.003
- Filichkin SA, Priest HD, Givan SA, Shen R, et al. (2010) Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Research. 20 (1): 45-58. https://doi.org/10.1101/gr.093302.109
- Garg R, Jain M (2013) RNA-Seq for transcriptome analysis in non-model plants. Methods Mol Biol 1069: 43-58. https://doi.org/10.1007/978-1-62703-613-9_4
- Helm BR, Rinehart JP, Yocum GD, Greenlee KJ, et al. (2017) Metamorphosis is induced by food absence rather than a critical weight in the solitary bee, Osmia lignaria. Proc Natl Acad Sci USA 114: 10924–10929. https://doi.org/10.1073/pnas.1703008114
- Herranz R, Mateos J, Marco R (2005) Diversification and independent evolution of troponin C genes in insects. J Mol Evol. 60 (1): 31-44. https://doi.org/10.1007/s00239-004-0031-x
- Ichihashi Y, Mutuku JM, Yoshida S, Shirasu K (2015) Transcriptomics exposes the uniqueness of parasitic plants. Brief Funct Genomics 14 (4): 275-282. https://doi.org/10.1093/bfgp/elv001
- Jia FL, Zhang QL (1999) Insect metamorphosis and evolution. Entomol Knowledge. 36: 363-370.
- Jindra M, Palli SR, Riddiford LM (2013) The juvenile hormone signaling pathway in insect development. Annu Rev Entomol. 58 (1): 181-204. https://doi.org/10.1146/annurev-ento-120811-153700
- Kanehisa M, Goto S, Kawashima S, Okuno Y, et al. (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res. 32 (90001): D277-D280. https://doi.org/10.1093/nar/gkh063
- Kim D, Pertea G, Trapnell C, Pimentel H, et al. (2013) TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. https://doi.org/10.1186/gb-2013-14-4-r36
- Kržič U, Rybin V, Leonard KR, Linke WA, et al. (2010) Regulation of oscillatory contraction in insect flight muscle by troponin. J Mol Biol. 397: 110-118. https://doi.org/10.1016/j.jmb.2010.01.039
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. https://doi.org/10.1186/gb-2009-10-3-r25
- Lappalainen T, Sammeth M, Friedländer MR, Pa TH, et al. (2013) Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501: 506-511.
- Li LT, Zhu YB, Ma JF, Li ZY, et al. (2013) An analysis of the Athetis lepigone transcriptome from four developmental stages. PLoS One. 8: e73911. https://doi.org/10.1371/journal.pone.0073911
- Liu Z, Ling L, Xu J, Zeng B, et al. (2017) MicroRNA-14 regulates larval development time in Bombyx mori. Insect Biochem. Mol. Biol. 93: 57-65. https://doi.org/10.1016/j.ibmb.2017.12.009
- Malmstrom C (2012) Ecologists study the interactions of organisms and their environment. Nat Edu. Knowledge 3: 88.
- Malumbres M (2014) Cyclin-dependent kinases. Genome Biol. 15: 122. https://doi.org/10.1007/springerreference_173086
- Micciarelli AS. (1977) Effects of farnesyl methyl ether on embryos of Schistocerca Gregaria (Orthoptera). Acta Embryol Exp. 3: 295-303.
- Mortazavi A, Williams BA, Mccue K, Schaeffer L, et al. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 5 (7): 621-628. https://doi.org/10.1038/nmeth.1226
- Nishikawa H, Iijima T, Kajitani R, Yamaguchi J, et al. (2015) A genetic mechanism for female-limited batesian mimicry in Papilio butterfly. Nat Genet. 47 (4): 405-409. https://doi.org/10.1038/ng.3241
- Novák, VJA (1969) Morphogenetic analysis of the effects of juvenile hormone analogues and other morphogenetically active substances on embryos of Schistocerca gregaria (Forskål). J. Embryol. Exp. Morphol. 21: 1-21.
- Oppenheim SJ, Baker RH, Simon S, Desalle R (2015) We can't all be supermodels: the value of comparative transcriptomics to the study of non-model insects. Insect Mol. Biol. 24: 139-154. https://doi.org/10.1111/imb.12154
- Qi L, Fang Q, Zhao L, Xia H, et al. (2016) De novo assembly and developmental transcriptome analysis of the small white butterfly Pieris rapae. PLoS One. 11 (7): e0159258. https://doi.org/10.1371/journal.pone.0159258
- Resh, VH, Cardé, RT (2009) Encyclopedia of insects. (2nd edn). Academic Press. New York. https://doi.org/10.1016/b978-0-12-374144-8.x0001-x
- RiddifordRiddiford LM (2012) How does juvenile hormone control insect metamorphosis and reproduction? Gen. Comp. Endocrinol. 179 (3): 477-484. https://doi.org/10.1016/j.ygcen.2012.06.001
- Truman JW, Riddiford LM (1999) The origins of insect metamorphosis. Nature. 401: 457-452.
- Truman JW, Riddiford LM (2002) Endocrine insights into the evolution of metamorphosis in insects. Annu Rev Entomol 47: 467-500.
- Truman JW, Riddiford LM (2007) The morphostatic actions of juvenile hormone. Insect Biochem Mol Biol. 37: 761-760. https://doi.org/10.1016/j.ibmb.2007.05.011
- Wang P, Zhang X, Zhang J (2005) Molecular characterization of four midgut aminopeptidase N isozymes from the cabbage looper, Trichoplusia ni. Insect Biochem Mol Biol. 35: 611-620. https://doi.org/10.1016/j.ibmb.2005.02.002
- Wijesekera TP, Saurabh S, Dauwalder B (2016) Juvenile hormone is required in adult males for Drosophila courtship. PLoS One. 11: e0151912. https://doi.org/10.1371/journal.pone.0151912
- Xu ZF (2009) General entomology. Science Press, Beijing.
- Yang HJ, Zhou F, Sabhat A (2011) Expression pattern of enzymes related to juvenile hormone metabolism in the silkworm, Bombyx mori L. Mol Biol Rep. 38: 4337-4342. https://doi.org/10.1007/s11033-010-0559-3
- Zhang Y, Huang Q, Pennerman KK, Yu J, et al. (2016) Datasets for transcriptomic analyses of maize leaves in response to Asian corn borer feeding and/or jasmonic acid. Data in Brief. 7: 1010-1014. https://doi.org/10.1016/j.dib.2016.03.071
- Zhang YJ, Hao Y, Si F, Ren S, et al. (2014) The de novo transcriptome and its analysis in the worldwide vegetable pest, Delia antiqua (Diptera: Anthomyiidae). G3 (Bethesda) 4 (5): 851-859. https://doi.org/10.1534/g3.113.009779
- Zheng W, Peng T, He W, Zhang H (2012) High-throughput sequencing to reveal genes involved in reproduction and development in Bactrocera dorsalis (Diptera: Tephritidae). PLoS One. 7: e36463. https://doi.org/10.1371/journal.pone.0036463
- Zou Z, Lopez DL, Kanost MR, Evans JD, et al. (2006) Comparative analysis of serine protease‐related genes in the honey bee genome: possible involvement in embryonic development and innate immunity. Insect Mol Biol. 15: 603-614. https://doi.org/10.1111/j.1365-2583.2006.00684.x
Keywords:
Download:
Full PDF- Share This