The Disruption of Klf6-Related Super-Enhancer Induces Growth Inhibition and Apoptosis in Human HepG2 Cells
Received: January 14, 2018
Accepted: February 24, 2018
Published: February 28, 2018
Genet.Mol.Res. 17(1): gmr16039888
DOI: 10.4238/gmr16039888
Abstract
Klf6 is a member of the Krüppel-like factor family, which is a group of zinc finger transcription factors involved in differentiation and development. In general, Klf6 gene plays an important role in growth-related signal transduction pathways, cell proliferation, apoptosis, and angiogenesis. Recently, the Klf6 gene is considered as one of tumor suppressor genes in several kinds of tumours, but there is also a conflict of experimental data. In this study, we investigated the influence of Klf6-related super-enhancer (SE) on tumor cell growth in human hepatoma (HepG2) cells. As a result, SE-deleted clones were obtained via the CRISPR/Cas9-mediated gene-editing technique. Moreover, it was found that some crucial properties associated with malignancy in those clones were equally affected by the disruption of Klf6-related super-enhancer: proliferation, migration, clonogenic potential and drug resistance. These results contribute to the deeper understanding on the potential roles of the Klf6 related super enhancer in human hepatoma (HepG2) cells.
Introduction
Klf6 is a member of the Krüppel-like factor family, a group of zinc finger transcription factors involved in differentiation and development. It plays an important role in growth-related signal transduction pathways, cell proliferation, apoptosis, and angiogenesis [Jaya Sangodkar et al., 2009; Magali Humbert et al., 2011].
Although the exact mechanisms underlying the tumor suppressor roles of Klf6 are unknown, a number of relevant pathways have been described: transactivation of P21 WAF1 in a P53- independent manner, reduction of cyclin D1/cdk4 complexes via interaction with cyclin D1, inhibition of c-Jun proto-oncoprotein activities, decreased VEGF expression and induction of apoptosis [Benzeno S et al., 2004; SlavinDA et al., 2004; A.C. Racca et al., 2016].
Klf6 has been postulated as a tumor suppressor since its gene is frequently inactivated by loss of heterozygozity (LOH), somatic mutations and/or decreased expression in human cancers, but there are also conflicting experimental data [A. Difeo et al., 2009; Analisa DiFeo et al., 2009; Ceshi Chen et al., 2003].
Several independent reports described apoptotic or anti-apoptotic functions for Klf6. siRNA- mediated Klf6 knock down in hepatocarcinoma cell lines lead to a reduction in cell cycle progression and cells became more susceptible to DNA-damage induced apoptosis, indicating that Klf6 function is required for cell proliferation and survival in these cells [D.S. D’Astolfo et al., 2008; E. Sirach et al., 2007]. As a result, the Klf6 gene pathways are assumed to be very complex and diverse, and research urges clarification of the exact mechanism of Klf6, which plays an important role in tumorigenesis and development [Sigal Kremer-Tal et al., 2007; Ricardo C et al., 2011].
Recently, researchers have identified large non-coding enhancers, called super-enhancers, and attempted to identify their in vivo functions [Anna Vähärautio et al., 2014; Yanjun Wei et al., 2016]. Super-enhancers (SEs) shown to specifically regulate genes involved in cell identity and disease, including oncogenes [Adrienne R et al., 2015; Denes Hnisz et al., 2013; Denes Hnisz et al., 2015; Jakob Love´n et al., 2013; Marc R et al., 2014]. We used genome-editing techniques to identify Klf6-related super-enhancers in human HepG2 cells [Kumchol Ri et al., 2017; Cong L et al., 2013]. The results demonstrated that Klf6-related SE regulates the Klf6 gene expression through long-range DNA looping [Kumchol Ri et al., 2017]. Recent studies from our laboratory found that inhibition of the Klf6 gene expression by the CRISPR Technique inhibited proliferation and induced apoptosis in human hepatoma (HepG2) cells.
In this study, we obtained a clone of Klf6-related SE-deletion by using the CRISPR/Cas9- mediated genetic engineering technique [Kumchol Ri et al., 2017] and found that some crucial properties associated with malignancy in those clones, including proliferation, migration, clonogenic potential and drug resistance, were equally affected by the disruption of Klf6-related super-enhancer. These results suggest that the disruption of the Klf6-related SE induces growth inhibition and apoptosis in human HepG2 cells. These data lead us to explore and further define the potential role of the Klf6 related super enhancer in human hepatoma (HepG2) cells.
Materials and Methods
Cell culture and transfection experiments
Human HepG2 cells were purchased from the typical training content preservation committee cell bank, Chinese Academy of Sciences (China). The Cell culture and transfection experiment were performed as previously described (Kumchol Ri et al., 2017). The cells were passaged twice. For transfection, the cells were cultured with CRISPR plasmid and Lipofectamine 2000 (Addgene) for 6 hours (h).
After transfection, the cells were cultured for 48 h in fresh medium, 0.5 μg/mL of puromycin (Sigma) was supplemented to the cell culture medium for 3 days to select for the transfected cells. Cells were cultured for 24 h in a fresh medium, and clones were obtained by limiting dilution method, from which target clones were selected.
The target-specific CRISPR guide RNAs (sgRNAs) were designed using an online tool (http://crispr.mit.edu/). The designed oligonucleotides were inserted into pSpCas9 (BB)-2A-GFP (PX458) plasmid (Addgene) and used for cell transfection experiments.
Real-time quantitative PCR and Western blot assay
Total RNA was isolated from the target clones using TRIzol (Life Tech) extraction method. Primer sequences for the Klf6 genes and beta-actin are as follows: 5′-CGGACGCACAC AGGAGAAAA-3′/5′-CGGTGTGCTTTCGGAAGTG-3′ (Klf6) and 5′-GTGTGGCGACATATG CAGCT-3′/5′-CAAGATCAGCAGTCTCATTC-3′ (beta-actin). A total of 40 cycles were performed, each including 95°C for 30 seconds, 54°C for 30 seconds, and 72°C for 30 seconds. Assays were done in triplicate and Relative levels of gene expression were normalized to beta-actin.
Western blot assays were performed as previously described [Kumchol Ri et al., 2017]. Cells were re-suspended in lysis buffer. Proteins were separated on 10–15% SDS–PAGE gels. The following antibodies were used: rabbit polyclonal antibody to Klf6 (R-173): anti-human E-cadherin (CD324) antibody: goat polyclonal to beta-actin (I-19).
MTT assays and colony formation assay
MTT assay was performed as previously described [Sigal Kremer-Tal et al., 2007]. In brief, 200 μL of MTT (Sigma) in phosphate buffered saline (2 mg/mL) was added to each well. After 4 h incubation at 37â�?¦C, the supernatant was discarded, and the precipitate was dissolved in 1 mL of dimethyl sulfoxide. Plates were then read on a microplate reader at 490 nm.
Colony formation assay was preceded as previously described [Jaya Sangodkar et al., 2009]. A total of 100 cells were seeded in 6-well plates, and colonies consisting of at least 50 cells were counted after 14 days.
Migration assay
Migration ability was measured as previously described [Sigal Kremer-Tal et al., 2007]. Briefly, each clone cells was suspended at a concentration of 5 × 105cells/μL, and 3 × 104 cells were placed in each well. The wells were removed gently after a 24 hour incubation. The width of the scratch was measured at the beginning and every 12 hours during cell migration and quantified the wound closure rate as previously described [Sigal Kremer-Tal et al., 2007].
Cell cycle analysis
Each clone cells were trypsinized and collected, respectively. Cell pellets were fixed in 70% ethanol and stored at −20 °C overnight. Fixed cells were washed with PBS and resuspended in 50 mg/mL propidium iodide containing 125 U/mL RNase A. After 30 min incubation, DNA contents were analyzed by flow cytometry. The cells percentage in different phases of cell cycle was determined with the ModFit 2.0 computer program (Verity Software).
Detection of apoptotic cells
Cells were trypsinized and resuspended in a solution containing Annexin-V APC (Invitrogen) and SYTOX green (Invitrogen). Labelling intensities were quantified using Cell Quest software on a FacScalibur station.
Measurement of cell viability
Each clone cells were seeded at 5 ×103 cells/well of a 96-well dish. After 48h incubation, cells were treated with Gemcitabine at the different concentrations. After 48 h incubation, cell viability was measured using Cell titer96® Aqueous One Solution Cell Proliferation assay (MTS) (Promega).
Statistical analysis
Statistical analyses were performed using GraphPad Prism (GraphPad Software). Data represent the mean ± SEM or SD, as indicated. Differences between the means were analyzed using the 2-tailed unpaired Student’s t test or ANOVA or the Mann-Whitney U or Kruskal-Wallis test. A P value of less than 0.05 was considered significant.
Results
Disruption of Klf6-related SE regulates Klf6 expression in HepG2 cells
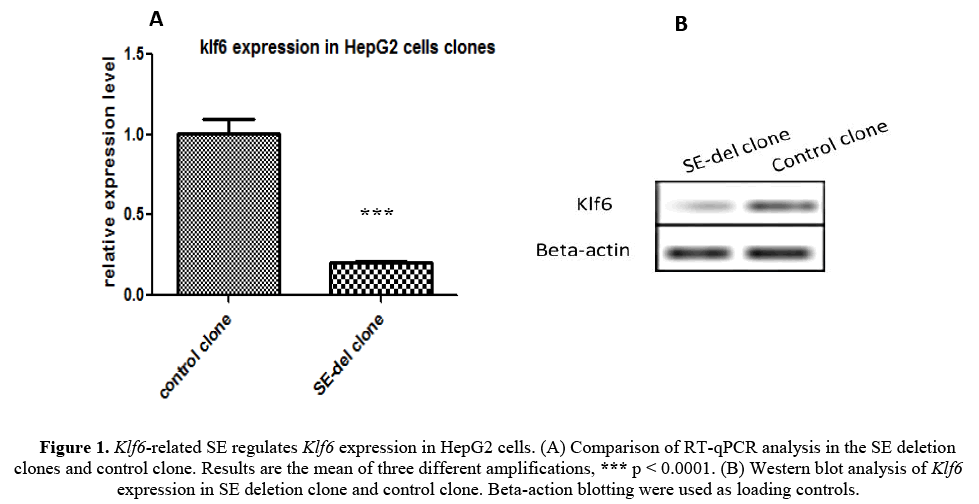
We first generated a clone of Klf6-related SE-deletion (SE-del) in in HepG2 cells using the CRISPR/Cas9 system to evaluate the functions of Klf6 gene (described in detail [Kumchol Ri et al., 2017]). RT-qPCR analysis was performed to examine the gene expression of Klf6 on the obtained SE-del clone (Figure 1A). As shown in the figure, the expression of Klf6 in the SE-del clone is reduced by 80% or more compared to the control.
Figure 1: Klf6-related SE regulates Klf6 expression in HepG2 cells. (A) Comparison of RT-qPCR analysis in the SE deletion clones and control clone. Results are the mean of three different amplifications, *** p < 0.0001. (B) Western blot analysis of Klf6 expression in SE deletion clone and control clone. Beta-action blotting were used as loading controls.
To further confirm the expression characteristics of Klf6, the Klf6 protein expression was tested using western blot analysis (Figure 1B). The Klf6 Protein expression also showed a significant decrease in SE-del clone compared to the control clone.
The results show that the Klf6-related SE plays an important role in the regulation of the expression of the Klf6 gene and that the disruption of the Klf6-related SE decreases the expression of the Klf6 gene by more than 80%. We next conducted an experiment to determine whether the inhibition of Klf6 gene expression had any biological relationship with some tumor properties.
Disruption of Klf6-related SE decreases clonogenic potential of HepG2 cells
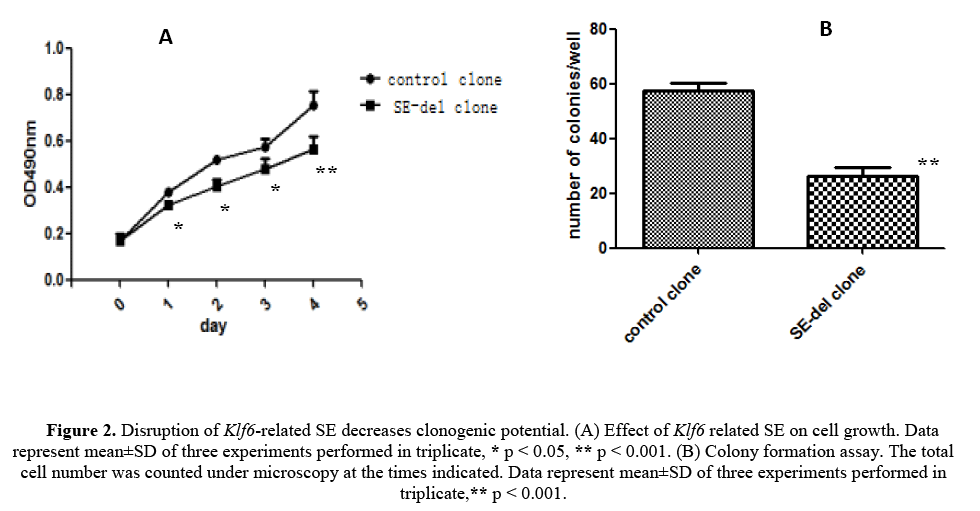
We have evaluated the effect of disruption of the Klf6-related ES on cell growth and clonogenic potential of the HepG2 cells, starting from the fact that malignant tumor cells exhibit a high clonogenic potential (Figures 2A and 2B). The results revealed strong inhibition of cell growth, but also remarkable inhibition of clonogenic potential in in SE-del clone, compared with the control clone. Therefore, it concluded that disruption of Klf6-related SE decreases clonogenic potential of HepG2 cells.
Figure 2: Disruption of Klf6-related SE decreases clonogenic potential. (A) Effect of Klf6 related SE on cell growth. Data represent mean±SD of three experiments performed in triplicate, * p < 0.05, ** p < 0.001. (B) Colony formation assay. The total cell number was counted under microscopy at the times indicated. Data represent mean±SD of three experiments performed in triplicate,** p < 0.001.
Disruption of Klf6-related SE decreases migratory potential
In general, tumor-associated genes can control cell migration in cancer. Therefore, wound-healing assays were performed to examine the effect of Klf6 related SE on HepG2 cells migration.
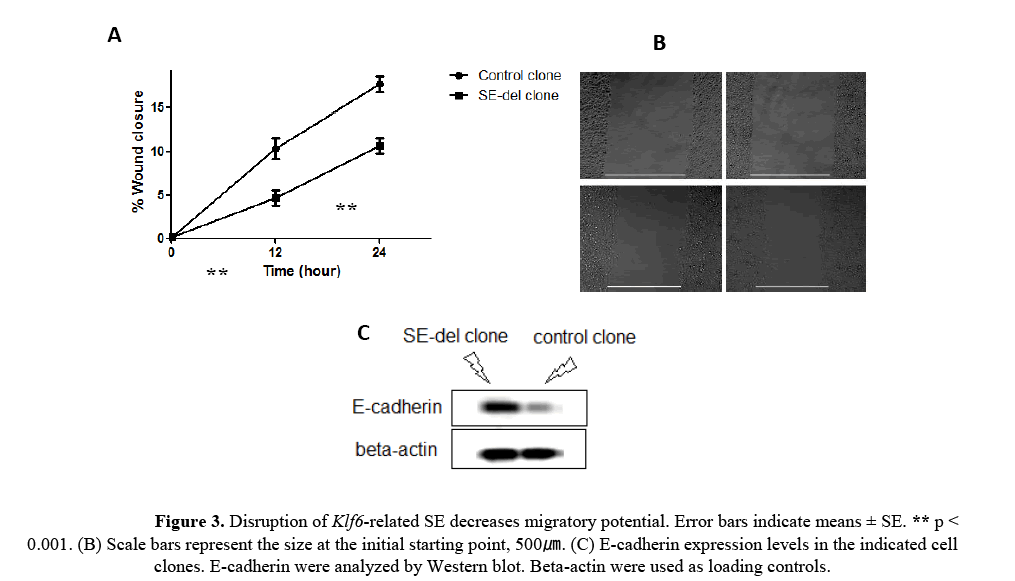
As shown in the Figures 3A and 3B, migration was decreased by 40-50% in SE-del clone, at 24h, compared with the control clone. The reduction of E-cadherin expression in cancer cells leads to epithelial- mesenchymal transition (EMT) [Hao L et al., 2012]. It also found that SE-del clone exhibited increased E-cadherin expression (Figure 3C). These results indicate that Klf6-related SE is involved in the regulation of migration capacity in HepG2 cells.
Figure 3: Disruption of Klf6-related SE decreases migratory potential. Error bars indicate means ± SE. ** p < 0.001. (B) Scale bars represent the size at the initial starting point, 500ã�?�?. (C) E-cadherin expression levels in the indicated cell clones. E-cadherin were analyzed by Western blot. Beta-actin were used as loading controls.
Disruption of Klf6-related SE induces changes in cell-cycle distribution
To determine the mechanisms of the reduced rate of cellular proliferation, we then conducted cell cycle analysis of SE-del clone and control clone using flow cytometry.
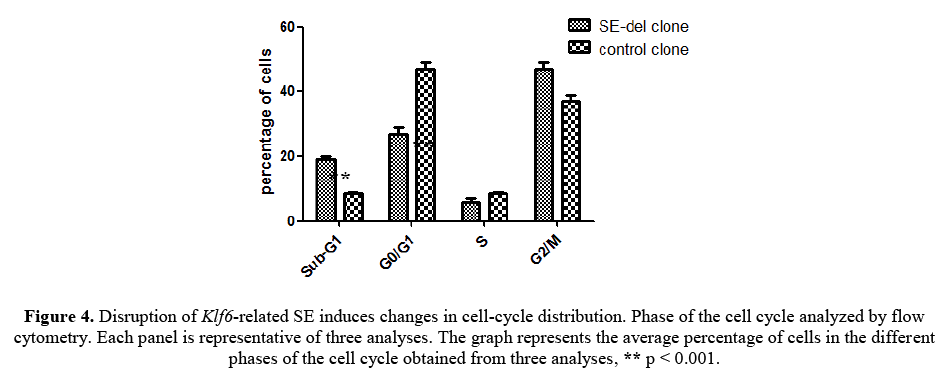
As shown in Figure 4, this reduced proliferation was accompanied by an increased proportion of sub-G1 phase and a lower percentage in Go/G1 phase in SE-del clone. SE-del clone increased the percentage of the total cell population in the sub-G1 phase, decreased the percentage of cells in the Go/G1 phase, but, had no consistent effect on the percentage of cells in the S phase, compared to the controls. Significantly, these data clearly demonstrated that ddisruption of Klf6-related SE increased the sub-G1 apoptotic cell population and decreased the Go/G1 cell population. These results increase the possibility that inhibition of cell proliferation induced by ddisruption of Klf6-related SE is associated with cell apoptosis.
Figure 4: Disruption of Klf6-related SE induces changes in cell-cycle distribution. Phase of the cell cycle analyzed by flow cytometry. Each panel is representative of three analyses. The graph represents the average percentage of cells in the different phases of the cell cycle obtained from three analyses, ** p < 0.001.
Disruption of Klf6-related SE induces apoptosis and increases sensitivity to Gemcitabine
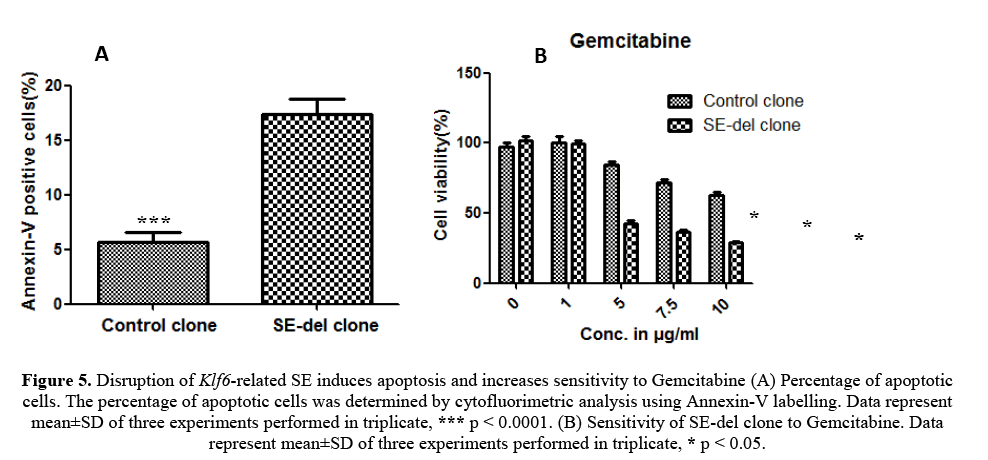
To determine the reduced rate of cellular proliferation, we also performed apoptosis analysis of SE-del clone and control clone using flow cytometry (Figure 5A). The increase of SE-del clone suggested a higher apoptotic rate, which was confirmed by Annexin V measurements. Altogether, these findings are in favour of the anti-proliferative and apoptotic effects of the SE-del clone.
Figure 5: Disruption of Klf6-related SE induces apoptosis and increases sensitivity to Gemcitabine (A) Percentage of apoptotic cells. The percentage of apoptotic cells was determined by cytofluorimetric analysis using Annexin-V labelling. Data represent mean±SD of three experiments performed in triplicate, *** p < 0.0001. (B) Sensitivity of SE-del clone to Gemcitabine. Data represent mean±SD of three experiments performed in triplicate, * p < 0.05.
The SE-del clone and control clone were grown in the presence of gemcitabine. After 48 h, cell viability was measured using an MTS assay. The graphs represent the average of three experiments in triplicate and are expressed as the percentage of viable cells relative to this percentage in the corresponding untreated cells (100%). The results observed that SE-del clone increased sensitivity to gemcitabine treatment (Figure 5B).
Although the molecular mechanisms remain to be elucidated, these findings demonstrate that Klf6-related SE increased sensitivity to the apoptotic and cytotoxic agent, gemcitabine.
Discussion
Clarifying the interactions between transcription factors and cancer properties is of great interest in understanding how cancer cells promote formation, development and metastasis through regulation of gene expression. Klf6 is a transcription factors involved in growth-related signal transduction pathways, cell proliferation, apoptosis, and angiogenesis. Klf6 has been postulated as a tumor suppressor since its gene is frequently inactivated by loss of heterozygozity (LOH), somatic mutations and/or decreased expression in human cancers [A. Difeo et al., 2009]. But several reports described apoptotic or anti-apoptotic functions for Klf6. siRNA-mediated Klf6 knock down in hepatocarcinoma cell lines lead to a reduction in cell cycle progression and cells became more susceptible to DNA-damage induced apoptosis, indicating that Klf6 function is required for cell proliferation and survival in these cells [D.S. D’Astolfo et al., 2008; E. Sirach et al., 2007]. Therefore, the exact functions of Klf6 gene remain controversial. This study used the CRISPR/Cas9 system to establish SE-del clone of Klf6-related super-enhancer and examined the function of Klf6-related SE in HepG2 cells. We previously used the CRISPR/Cas9 system to identify a transcription factor, Klf6-related SE, in HepG2 cells [Kumchol Ri et al., 2017]. During these experiments, it found that disruption of Klf6-related SE inhibits cell proliferation and induces apoptosis in in HepG2 cells.
The SE-del clones exhibited decreased Klf6 gene expression and endogenous Klf6 protein expression compared to the control clones (Figure 1). These results demonstrate that the Klf6-related SE regulates the expression of Klf6 gene in in HepG2 cells.
In our study, disruption of the Klf6-related SE alters cell growth, but also significantly decreased malignant potential, including colony formation (Figure 2), migration (Figure 3), changes in cell-cycle distribution (Figure 4) and drug resistance (Figure 5). These results suggest that the Klf6-related SE activates cancer cell properties. These data are consistent with literatures [D.S. D’Astolfo et al., 2008; E. Sirach et al., 2007] described apoptotic or anti-apoptotic functions for Klf6.
Our results demonstrated that the Klf6-related SE activates migration capacity via E-cadherin expression. E-cadherin promotes epithelial cell-cell adhesions, and decreased E-cadherin expression is important for EMT [Hao L et al., 2012].
Our data indicate that the Klf6-related SE is relevant to drug resistance. These results indicate that the Klf6-related SE is involved in the tumorigenic potential of HepG2 cells which is consistent with these previous studies [Benzeno S et al., 2004; E. Sirach et al., 2007; D.S. D’Astolfo et al., 2008].
Conclusion
To sum up, we established the SE-del clone, in which the Klf6-related super-enhancer was deleted via the CRISPR/Cas9 system. Our results indicate that the Klf6-related super-enhancer activates many properties associated with malignancy in HepG2 cells, including colony formation capacity, migration capability, and drug resistance. However, it is necessary to further explore and define the potential role of the Klf6-related super-enhancer transcriptional network in the regulation of Klf6 signaling in human hepatoma (HepG2) cells.
Acknowledgment
This work was supported by the National Natural Science Foundation of Korea (Grant No. 54289562).
Conflicts of interest
The authors declare that they have no conflict of interest.
About the Authors
Corresponding Author
KumChol Ri
School of Management, Harbin Institute of Technology, China
- Email:
- 15124579835@163.com
References
- Difeo A, Huang F, Sangodkar J, Terzo EA, et al. (2009). Klf6-SV1 is a novel antiapoptotic protein that targets the BH3-only protein NOXA for degradation and whose inhibition extends survival in an ovarian cancer model. Cancer Res. 69: 4733-4741. https://doi.org/10.1158/0008-5472.can-08-4282
- Racca AC, Ridano ME, Bandeira CL, Bevilacqua E, et al. (2016). Low oxygen tension induces Krüppel-Like Factor 6 expression in trophoblast cells. Placenta. 45: 50-57. https://doi.org/10.1016/j.placenta.2016.07.006
- Adrienne R, Niederriter, Arushi Varshney, Stephen C, et al. (2015). Super enhancers in cancers, complex disease, and developmental disorders. Genes. 6: 1183-1200. https://doi.org/10.3390/genes6041183
- Analisa DiFeo, John A. Martignetti, Goutham Narla, (2009). The role of Klf6 and its splice variants in cancer therapy. Drug Resistance Updates. 12: 1-7. https://doi.org/10.1016/j.drup.2008.11.001
- Anna Vähärautio, Jussi Taipale (2014). Cancer by super-enhancer. Science, 346 (6215), 1291-1292.
- Benzeno S, Narla G, Allina J, Cheng GZ, et al. (2004). Cyclin-dependent kinase inhibition by the Klf6 tumor suppressor protein through interaction with cyclin D1. Cancer Res. 64:3885-3891. https://doi.org/10.1158/0008-5472.can-03-2818
- Ceshi Chen, Eija-Riitta Hyytinen, Xiaodong Sun, Helin HJ, et al. (2003). Deletion, mutation, and loss of expression of Klf6 in human prostate cancer. American Journal of Pathology. 162: 1349-1354. https://doi.org/10.1016/s0002-9440 (10)63930-2
- Cong L, Ran FA, Cox D, Lin S, et al. Multiplex genome engineering using CRISPR/Cas systems. Science, 339: 819-823 (2013).
- D’Astolfo DS, Gehrau RC, Bocco JL, Koritschoner NP, et al. Silencing of the transcription factor Klf6 by siRNA leads to cell cycle arrest and sensitizes cells to apoptosis induced by DNA damage. Cell Death Differ. 15 (2008) 613-616. https://doi.org/10.1038/sj.cdd.4402299
- Denes Hnisz, Brian J. Abraham, Tong Ihn Lee, Ashley Lau, et al. (2013). Young, super-enhancers in the control of cell identity and disease. Cell. 155: 934-947.
- Denes Hnisz, Jurian Schuijers, Charles Y Lin, Abraham S Young, et al. (2015). Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Molecular Cell. 5: 1-9. https://doi.org/10.1016/j.molcel.2015.02.014
- Sirach E, Bureau C, Peron JM, Pradayrol L, et al. (2007). Klf6 transcription factor protects hepatocellular carcinoma-derived cells from apoptosis. Cell Death Differ. 14:1202-1210. https://doi.org/10.1038/sj.cdd.4402114
- Hao L, Ha JR, Kuzel P, Garcia E, et al. (2012). Cadherin switch from E-to N-cadherin in melanoma progression is regulated by the PI3K/PTEN pathway through Twist and Snail. Br J Dermatol. 166:1184-1197. https://doi.org/10.1111/j.1365-2133.2012.10824.x
- Jakob Love´n, Heather A. Hoke, Charles Y. Lin, Ashley Lau, et al. (2013). Selective inhibition of tumor oncogenes by disruption of super enhancers. Cell. 153: 320-334.
- Jaya Sangodkar, Analisa DiFeo, Lauren Feld, Romina Bromberg, et al. (2009). Targeted reduction of Klf6-SV1 restores chemotherapy sensitivity in resistant lung adenocarcinoma. Lung Cancer. 66: 292-297. https://doi.org/10.1016/j.lungcan.2009.02.014
- Jaya Sangodkar, Jiayan Shi, Analisa DiFeo, Rachel Schwartz, et al. (2009). Functional role of the Klf6 tumour suppressor gene in gastric cancer. European Journal of Cancer. 45: 666-676. https://doi.org/10.1016/j.ejca.2008.11.009
- Kum Chol Ri, Jong Su Kim, Chol Kim. (2017). Identification of Klf6-related super enhancer in human hepatoma (HepG2) cells by CRISPR technique. Genetics and Molecular Research 16 (4): gmr16039841. https://doi.org/10.4238/gmr16039841
- Magali Humbert, Veronika Halter, Deborah Shan, Judith Laedrach, et al. (2011). Deregulated expression of Kruppel-like factors in acute myeloid leukemia. Leukemia Research, 35: 909-913. https://doi.org/10.1016/j.leukres.2011.03.010
- Marc R, Mansour, Abraham BJ, Lars Anders, et al. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 346: 6215.
- Ricardo C. Gehrau, Diego S. D’Astolfo, Verónica Andreoli, et al. (2011). Differential expression of the Klf6 tumor suppressor gene upon cell damaging treatments in cancer cells. Mutation Research. 707: 15-23. https://doi.org/10.1016/j.mrfmmm.2010.12.002
- Sigal Kremer-Tal, Goutham Narla, Yingbei Chen, Eldad Hod, et al. (2007). Downregulation of Klf6 is an early event in hepatocarcinogenesis and stimulates proliferation while reducing differentiation. Journal of Hepatology. 46: 645-654. https://doi.org/10.1016/j.jhep.2006.10.012
- Slavin DA, Koritschoner NP, Prieto CC, Lopez-Diaz FJ, et al. (2004). A new role for the kruppel-like transcription factor Klf6 as an inhibitor of c-jun proto-oncoprotein function. Oncogene. 23:8196-8205. https://doi.org/10.1038/sj.onc.1208020
- Yanjun Wei, Shumei Zhang, Shipeng Shang, Bin Zhang, et al. (2016). SEA: A super-enhancer archive. Nucleic Acids Research. 172-179.
Keywords:
Download:
Full PDF- Share This